 |
Code Documentation 3.1
Social Network Visualizer
|
 |
Code Documentation 3.1
Social Network Visualizer
|
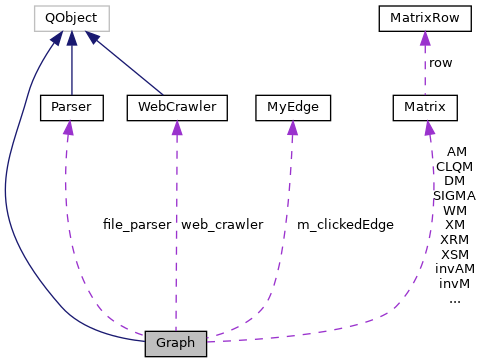
The Graph class This is the main class for a Graph, used in conjuction with GraphVertex, Parser and Matrix objects. Graph class methods are the interface to various analysis algorithms GraphVertex class holds each vertex data (colors, strings, statistics, etc) Matrix class holds the adjacency matrix of the network. Parser class loads files of networks. More...
#include <graph.h>
Public Types | |
| enum | ModStatus { NewNet = -1 , SavedUnchanged = 0 , MinorOptions = 1 , VertexMetadata = 2 , EdgeMetadata = 3 , VertexPositions = 4 , MajorChanges = 10 , VertexCount = 11 , EdgeCount = 12 , VertexEdgeCount = 13 } |
| enum | Clustering { Single_Linkage = 0 , Complete_Linkage = 1 , Average_Linkage = 2 } |
Public Slots | |
| int | relationCurrent () |
| Gets the current relation number. | |
| QString | relationCurrentName () const |
| Gets the current relation name. | |
| void | relationCurrentRename (const QString &newName) |
| Overload. Renames current relation to newName, without emitting any signal to MW. | |
| void | relationCurrentRename (const QString &newName, const bool &signalMW) |
| Renames current relation to newName, optionally emitting a signal to MW. | |
| void | vertexCreate (const int &number, const int &size, const QString &color, const QString &numColor, const int &numSize, const QString &label, const QString &labelColor, const int &labelSize, const QPointF &p, const QString &shape, const QString &iconPath=QString(), const bool &signalMW=false) |
| Creates a new vertex. | |
| void | graphFileLoaded (const int &fileType, const QString &fName=QString(), const QString &netName=QString(), const int &totalNodes=0, const int &totalLinks=0, const int &edgeDirType=0, const qint64 &elapsedTime=0, const QString &message=QString()) |
| Stores loaded file name, graph name, sets edge direction type and signals MW to update the UI. | |
| void | vertexRemoveDummyNode (int) |
| Deletes a dummy node. | |
| void | graphLoadedTerminateParserThreads (QString reason) |
| Graph::graphLoadedTerminateParserThreads. | |
| void | setSelectionChanged (const QList< int > selectedVertices, const QList< SelectedEdge > selectedEdges) |
| Sets the user-selected vertices and edges. | |
| void | graphClickedEmptySpace (const QPointF &p) |
| Resets the clicked edge and node. | |
| bool | edgeCreate (const int &v1, const int &v2, const qreal &weight, const QString &color, const int &type=0, const bool &drawArrows=true, const bool &bezier=false, const QString &label=QString(), const bool &signalMW=true) |
| Checks a) if edge exists and b) if the reverse edge exists Calls edgeAdd to add the new edge to the Graph, then emits drawEdge() which calls GW::drawEdge() to draw the new edge. Called from homonymous signal of Parser class. Also called from MW when user clicks on the "add link" button Also called (via MW) from GW when user middle-clicks on two nodes. | |
| void | edgeCreateWebCrawler (const int &source, const int &target) |
| Called from WebCrawler when it finds an new link Calls edgeCreate() method with initEdgeColor. | |
| void | vertexCreateAtPos (const QPointF &p) |
| Creates a new vertex in the given position. | |
| void | vertexCreateAtPosRandom (const bool &signalMW=false) |
| Creates a new randomly positioned vertex with default values. | |
| void | vertexCreateAtPosRandomWithLabel (const int &i, const QString &label, const bool &signalMW=false) |
| Creates a new randomly positioned vertex with specific number and label. All other values are from the defaults. | |
| void | relationSet (int relNum=RAND_MAX, const bool &updateUI=true) |
| Changes the current relation, and optionally emits signals to MW/GW (default: true) | |
| void | relationNext () |
| Changes graph to next relation. | |
| void | relationPrev () |
| Changes graph to previous relation. | |
| void | canvasSizeSet (const int &width, const int &height) |
| Sets the size of the canvas. | |
| double | canvasMaxRadius () const |
| Gets the max radius of the canvas. | |
| qreal | canvasMinDimension () const |
| Gets the min dimensions of the canvas. | |
| double | canvasVisibleX (const double &x) const |
| Checks if x is visible inside the canvas usable area and if not returns an adjusted x-coordinate. | |
| double | canvasVisibleY (const double &y) const |
| Checks if y is visible inside the canvas usable area and if not returns an adjusted y-coordinate. | |
| double | canvasRandomX () const |
| Returns a random x-coordinate adjusted to be visible inside the canvas usable area. | |
| double | canvasRandomY () const |
| Returns a random y-coordinate adjusted to be visible inside the canvas usable area. | |
| void | vertexIsolatedAllToggle (const bool &toggle) |
| Toggles the status of all isolated vertices (thos without links) | |
| void | vertexClickedSet (const int &v, const QPointF &p) |
| Sets the clicked vertex. | |
| void | edgeClickedSet (const int &v1, const int &v2, const bool &openMenu=false) |
| Sets the clicked edge. | |
| void | edgeFilterByWeight (const qreal, const bool) |
| Filters (disables) edges according the specified threshold weight. | |
| void | edgeFilterByRelation (int relation, bool status) |
| Toggles (enables or disables) all edges of the given relation. | |
| void | edgeFilterUnilateral (const bool &toggle) |
| Enables or disables unilateral edges in current relationship. | |
| void | startWebCrawler (const QUrl &startUrl, const QStringList &urlPatternsIncluded, const QStringList &urlPatternsExcluded, const QStringList &linkClasses, const int &maxNodes, const int &maxLinksPerPage, const bool &intLinks, const bool &childLinks, const bool &parentLinks, const bool &selfLinks, const bool &extLinksIncluded, const bool &extLinksCrawl, const bool &socialLinks, const bool &delayedRequests) |
| Creates a new WebCrawler, that will parse the downloaded HTML code of each webpage we download. Moves the WebCrawler to a new thread and starts the thread. Then creates the fist node (initial url), and starts the web spider to download the first HTML page. Called by MW with user options. | |
| void | slotHandleCrawlerRequestReply () |
| Gets the reply of a MW network request made by Web Crawler, and emits that reply as is to the Web Crawler. | |
| void | webSpider () |
| A loop, that takes urls awaiting in front of the urlQueue, and signals to the MW to make the network request. | |
| QString | htmlEscaped (QString str) const |
| Helper method, returns a nice qstring where all html special chars are encoded. | |
Signals | |
| void | signalWebCrawlParse (QNetworkReply *reply) |
| void | signalNetworkManagerRequest (const QUrl ¤tUrl, const NetworkRequestType &type) |
| void | signalProgressBoxCreate (const int max=0, const QString msg="Please wait") |
| void | signalProgressBoxKill (const int max=0) |
| void | signalProgressBoxUpdate (const int &count=0) |
| void | signalGraphSavedStatus (const int &status) |
| void | signalGraphModified (const bool &undirected, const int &vertices, const int &edges, const qreal &density, const bool ¬Saved=true) |
| void | signalGraphLoaded (const int &fileType, const QString &fileName=QString(), const QString &netName=QString(), const int &totalNodes=0, const int &totalLinks=0, const qreal &density=0, const qint64 &elapsedTime=0, const QString &message=QString()) |
| void | statusMessage (const QString &message) |
| void | signalDatasetDescription (QString) |
| void | signalNodeClickedInfo (const int &number=0, const QPointF &p=QPointF(), const QString &label=QString(), const int &inDegree=0, const int &outDegree=0) |
| void | signalEdgeClicked (const MyEdge &edge=MyEdge(), const bool &openMenu=false) |
| void | signalRelationAddToMW (const QString &newRelation) |
| void | signalRelationsClear () |
| void | signalRelationRenamedToMW (const QString &newRelName) |
| void | signalRelationChangedToGW (int) |
| void | signalRelationChangedToMW (const int &relIndex=RAND_MAX) |
| void | signalSelectionChanged (const int &selectedVertices, const int &selectedEdges) |
| void | signalPromininenceDistributionChartUpdate (QAbstractSeries *series, QAbstractAxis *axisX=Q_NULLPTR, const qreal &min=0, const qreal &max=0, QAbstractAxis *axisY=Q_NULLPTR, const qreal &minF=0, const qreal &maxF=0) |
| void | signalDrawNode (const QPointF &p, const int &num, const int &size, const QString &nodeShape, const QString &nodeIconPath, const QString &nodeColor, const QString &numberColor, const int &numSize, const int &numDistance, const QString &label, const QString &labelColor, const int &labelSize, const int &labelDistance) |
| void | signalRemoveNode (int) |
| void | signalDrawEdge (const int &v1, const int &v2, const qreal &weight, const QString &label="", const QString &color="black", const int &type=0, const bool &drawArrows=true, const bool &bezier=false, const bool &weightNumbers=false) |
| void | signalRemoveEdge (const int &v1, const int &v2, const bool &removeReverse) |
| void | signalSetEdgeVisibility (const int &relation, const int &source, const int &target, const bool &toggle, const bool &preserveReverseEdge=false, const int &edgeWeight=1, const int &reverseEdgeWeight=1) |
| void | setVertexVisibility (const int &number, const bool &toggle) |
| void | setNodePos (const int &, const qreal &, const qreal &) |
| void | signalNodesFound (const QList< int > foundList) |
| void | setNodeSize (const int &v, const int &size) |
| void | setNodeShape (const int &v, const QString &shape, const QString &iconPath=QString()) |
| void | setNodeColor (const int &v, const QString &color) |
| void | setNodeLabel (const int &v, const QString &label) |
| void | setNodeNumberColor (const int &v, const QString &color) |
| void | setNodeNumberSize (const int &v, const int &size) |
| void | setNodeNumberDistance (const int &v, const int &distance) |
| void | setNodeLabelSize (const int &v, const int &size) |
| void | setNodeLabelColor (const int &v, const QString &color) |
| void | setNodeLabelDistance (const int &v, const int &distance) |
| void | setEdgeWeight (const int &v1, const int &v2, const qreal &weight) |
| void | signalEdgeType (const int &v1, const int &v2, const int &type) |
| void | setEdgeColor (const int &v1, const int &v2, const QString &color) |
| void | setEdgeLabel (const int &v1, const int &v2, const QString &label) |
| void | addGuideCircle (const double &, const double &, const double &) |
| void | addGuideHLine (const double &y0) |
Public Member Functions | |
| Graph (const int &reserveVerticesSize=5000, const int &reserveEdgesPerVertexSize=500) | |
| Constructs a Graph. | |
| ~Graph () | |
| Destroys the Graph object. | |
| void | clear (const QString &reason="") |
| Clears all vertices. | |
| QString | getFileName () const |
| Returns the file name of the current graph, if any. | |
| void | setFileName (const QString &fileName) |
| Sets the file name of the current graph. | |
| QString | getName () const |
| Returns the name of the current graph. | |
| void | setName (const QString &graphName) |
| Sets the name of the current graph. | |
| bool | isSaved () const |
| Returns true if the graph is saved. | |
| bool | isLoaded () const |
| Returns true if a graph has been loaded from a file. | |
| int | getFileFormat () const |
| Returns the format of the last file opened. | |
| void | setFileFormat (const int &fileFormat) |
| bool | isFileFormatExportSupported (const int &fileFormat) const |
| Returns true if the fileFormat is supported for saving. | |
| void | setModStatus (const int &graphNewStatus, const bool &signalMW=true) |
| Sets the graph modification status. | |
| bool | isModified () const |
| Returns true of graph is modified (edges/vertices added/removed) | |
| void | loadFile (const QString fileName, const QString codecName, const int format, const QString delimiter=QString(), const int sm_two_mode=1, const bool sm_has_labels=false) |
| Loads a graph from a given file. | |
| void | saveToFile (const QString &fileName, const int &fileType, const bool &saveEdgeWeights=true, const bool &saveZeroWeightEdges=false) |
| Saves the current graph to a file. | |
| bool | saveToPajekFormat (const QString &fileName, QString networkName="", int maxWidth=0, int maxHeight=0) |
| Saves the active graph to a Pajek-formatted file. | |
| bool | saveToAdjacencyFormat (const QString &fileName, const bool &saveEdgeWeights=true) |
| Saves the active graph to an adjacency-formatted file. | |
| bool | saveToGraphMLFormat (const QString &fileName, const bool &saveZeroWeightEdges=false, QString networkName="", int maxWidth=0, int maxHeight=0) |
| Saves the current graph to a GraphML-formatted file. | |
| bool | saveToDotFormat (QString fileName) |
| TODO Saves the active graph to a UCINET-formatted file. | |
| QString | graphMatrixTypeToString (const int &matrixType) const |
| Helper method, return the human readable name of matrix type. | |
| int | graphMatrixStrToType (const QString &matrix) const |
| Helper method, return the matrix type of human readable matrix name . | |
| QString | graphMetricTypeToString (const int &metricType) const |
| Helper method, return the human readable name of metric type. | |
| int | graphMetricStrToType (const QString &metricStr) const |
| Helper method, return the identifier of a metric. | |
| QString | graphClusteringMethodTypeToString (const int &methodType) const |
| Helper method, return the human readable name of clustering method type. | |
| int | graphClusteringMethodStrToType (const QString &method) const |
| Helper method, return clustering method type from the human readable name of it. | |
| int | relations () |
| Returns the count of relationships in this Graph. | |
| void | relationsClear () |
| Clears relationships in this Graph. | |
| void | relationAdd (const QString &relName, const bool &changeRelation=false) |
| Adds a new relation to the graph. | |
| int | vertexNumberMax () |
| Returns the number of the last vertex in the graph. | |
| int | vertexNumberMin () |
| Returns the number of the first vertex in the graph. | |
| int | vertexDegreeOut (int) |
| Returns the outDegree (sum of outbound edge weights) of vertex v1. | |
| int | vertexDegreeIn (int) |
| Returns the inDegree (sum of inbound edge weights) of vertex v1. | |
| QList< int > | vertexNeighborhoodList (const int &v1) |
| Returns a list of all vertices mutually connected to vertex v1 in the current relation. | |
| QSet< int > | vertexNeighborhoodSet (const int &v1) |
| Returns the set of all vertices mutually connected to vertex v1 in the current relation. | |
| bool | vertexIsolated (const int &v1) const |
| Checks if vertex is isolated. | |
| int | vertexExists (const int &v1) |
| Checks if the given vertex exists in the graph. | |
| int | vertexExists (const QString &label) |
| Checks if there is a vertex with a specific label exists in the graph. | |
| bool | vertexFindByNumber (const QStringList &numList) |
| Finds vertices in strList by their number. | |
| bool | vertexFindByLabel (const QStringList &labelList) |
| Finds vertices by their label. | |
| bool | vertexFindByIndexScore (const int &index, const QStringList &thresholds, const bool &considerWeights, const bool &inverseWeights=false, const bool &dropIsolates=false) |
| Finds vertices by their index score. | |
| void | vertexRemove (const int &v1) |
| Removes the vertex v1 from the graph First, it removes all edges to doomed from other vertices Then it changes the vpos of all subsequent vertices inside m_graph Finally, it removes the vertex. | |
| void | vertexSizeInit (const int) |
| Sets the initial vertex size. | |
| void | vertexSizeSet (const int &v, const int &newsize) |
| Changes the size of a vertex v or all vertices if v=0. | |
| int | vertexSize (const int &v) const |
| Returns the size of vertex v. | |
| void | vertexShapeSetDefault (const QString, const QString &iconPath=QString()) |
| Sets the default vertex shape and iconPath. | |
| void | vertexShapeSet (const int &v, const QString &shape, const QString &iconPath=QString()) |
| Changes the shape and iconPath of vertex v1, or all vertices if v1=-1. | |
| QString | vertexShape (const int &v) |
| Returns the shape of this vertex. | |
| QString | vertexShapeIconPath (const int &v) |
| Returns the IconPath of vertex v1. | |
| bool | graphHasVertexCustomIcons () const |
| void | vertexColorInit (const QString &color) |
| Graph::vertexColorInit default vertex color initialization. | |
| void | vertexColorSet (const int &v, const QString &color) |
| Changes the color of vertex v1. | |
| QColor | vertexColor (const int &v) const |
| Graph::vertexColor. | |
| void | vertexNumberColorInit (const QString &color) |
| Changes the initial color of the vertex numbers. | |
| void | vertexNumberColorSet (const int &v=0, const QString &color="#000000") |
| Graph::vertexColorSet Changes the color of vertex v1. | |
| void | vertexNumberSizeInit (const int &size) |
| Changes the initial size of vertex numbers. | |
| void | vertexNumberSizeSet (const int &v, const int &newsize) |
| Changes the size of vertex v number. | |
| void | vertexNumberDistanceInit (const int &distance) |
| Changes the initial distance of vertex numbers. | |
| void | vertexNumberDistanceSet (const int &v, const int &newDistance) |
| Changes the distance.of vertex v number from the vertex. | |
| void | vertexLabelSet (const int &v, const QString &label) |
| Changes the label of a vertex v1. | |
| QString | vertexLabel (const int &v) const |
| Returns the label of a vertex v1. | |
| void | vertexLabelsVisibilitySet (bool toggle) |
| void | vertexLabelSizeInit (int newSize) |
| Graph::vertexLabelSizeInit Changes the default size of vertex labels. | |
| void | vertexLabelSizeSet (const int &v, const int &labelSize) |
| Changes the label size of vertex v1 or all vertices if v1=0. | |
| void | vertexLabelColorInit (QString color) |
| Graph::vertexLabelColorInit Changes the default vertex label color. | |
| void | vertexLabelColorSet (const int &v1, const QString &color) |
| Changes the label color of vertex v1 or all vertices if v1 = 0. | |
| void | vertexLabelDistanceInit (const int &distance) |
| Changes the default distance of vertex labels. | |
| void | vertexLabelDistanceSet (const int &v, const int &newDistance) |
| Changes the distance.of vertex v label from the vertex. | |
| void | vertexLabelDistanceAllSet (const int &newDistance) |
| Changes the distance of all vertex labels from their vertices. | |
| void | vertexPosSet (const int &v, const int &x, const int &y) |
| Changes the position of the given vertex. | |
| QPointF | vertexPos (const int &v1) const |
| Returns the position of the given vertex. | |
| int | vertexClicked () const |
| Returns the number of the clicked vertex. | |
| int | vertices (const bool &dropIsolates=false, const bool &countAll=false, const bool &recount=false) |
| Gets the number of vertices in the graph. | |
| int | vertexEdgesOutbound (int i) |
| Returns the number of outbound edges (arcs) from vertex v1. | |
| int | vertexEdgesInbound (int i) |
| Returns the number of inbound edges (arcs) to vertex v1. | |
| int | verticesWithOutboundEdges () |
| Returns the sum of vertices having edgesOutbound. | |
| int | verticesWithInboundEdges () |
| Returns the sum of vertices having edgesInbound. | |
| int | verticesWithReciprocalEdges () |
| Returns the sum of vertices having reciprocal edges. | |
| QList< int > | verticesListIsolated () |
| Returns a list of all isolated vertices inside the graph. | |
| QList< int > | verticesList () |
| Returns a list of all vertices numbers inside the graph. | |
| QSet< int > | verticesSet () |
| Returns a QSet of all vertices numbers inside the graph. | |
| void | verticesCreateSubgraph (QList< int > vList, const int &type=SUBGRAPH_CLIQUE, const int ¢er=0) |
| Creates a subgraph (clique, star, cycle, line) with vertices in vList Iff vList is empty, then fallbacks to the m_verticesSelected. | |
| int | edgesEnabled () |
| Returns the number |E| of graph - only the enabled edges. | |
| MyEdge | edgeClicked () |
| Returns clicked edge. | |
| qreal | edgeExists (const int &v1, const int &v2, const bool &checkReciprocal=false) |
| Checks if there is an edge from v1 to v2 and returns the weight, if the edge exists. | |
| qreal | edgeExistsVirtual (const int &v1, const int &v2) |
| Checks if there is an edge from v1 to v2, even weight = 0 and returns the weight, if the edge exists or RAND_MAX if the edge does not exist at all. | |
| void | edgeOutboundStatusSet (const int &source, const int &target, const bool &toggle=false) |
| Toggles the status of outbound edge source -> target at source vertex. | |
| void | edgeInboundStatusSet (const int &target, const int &source, const bool &toggle=false) |
| Toggles the status of inbound edge target <- source at target vertex. | |
| void | edgeRemove (const int &v1, const int &v2, const bool &removeReverse=false) |
| Removes the directed arc v1->v2 or, if the graph is undirected, the edge v1 <->v2. | |
| void | edgeRemoveSelected (SelectedEdge &selectedEdge, const bool &removeReverse) |
| Removes a SelectedEdge. | |
| void | edgeRemoveSelectedAll () |
| Removes all selected edges. | |
| bool | edgeSymmetric (const int &v1, const int &v2) |
| Returns TRUE if edge(v1, v2) is symmetric, i.e. (v1,v2) == (v2,v1). | |
| void | edgeTypeSet (const int &v1, const int &v2, const qreal &w, const int &dirType=EdgeType::Directed) |
| Changes the direction type of an existing edge. | |
| void | edgeWeightSet (const int &v1, const int &v2, const qreal &w, const bool &undirected=false) |
| Changes the weight of the edge from vertex v1 to v2 (and optionally of the reverse edge) | |
| qreal | edgeWeight (const int &v1, const int &v2) const |
| Returns the weight of the edge v1->v2. | |
| void | edgeWeightNumbersVisibilitySet (const bool &toggle) |
| Changes the visibility of edge weight numbers. | |
| void | edgeLabelSet (const int &v1, const int &v2, const QString &label) |
| Changes the label of edge v1->v2. | |
| QString | edgeLabel (const int &v1, const int &v2) const |
| Returns the label of edge v1->v2. | |
| void | edgeLabelsVisibilitySet (const bool &toggle) |
| Toggles the visibility of edge labels. | |
| void | edgeColorInit (const QString &) |
| Saves the default edge color Used by random network creation methods. | |
| void | edgeColorSet (const int &v1, const int &v2, const QString &color) |
| Changes the color of edge v1->v2. | |
| QString | edgeColor (const int &v1, const int &v2) |
| Returns the color of the directed edge v1->v2. | |
| bool | edgeColorAllSet (const QString &color, const int &threshold=RAND_MAX) |
| Changes the color of all enabled edges. | |
| bool | isEmpty () const |
| Returns true if the current graph has no vertices at all. | |
| QList< int > | getSelectedVertices () const |
| Returns a QList of user-selected vertices. | |
| int | getSelectedVerticesCount () const |
| Returns count of user-selected vertices. | |
| int | getSelectedVerticesMin () const |
| Returns min of user-selected vertices. | |
| int | getSelectedVerticesMax () const |
| Returns max of user-selected vertices. | |
| QList< SelectedEdge > | getSelectedEdges () const |
| Returns a QList of user-selected edges in pair<int,int> | |
| int | getSelectedEdgesCount () const |
| Returns the count of user-selected edges. | |
| int | getGeodesicsCount () |
| Returns the number of geodesics (shortest-paths) in the graph. | |
| qreal | graphDensity () |
| Gets the graph density (if computed) or computes it again. | |
| bool | isWeighted () |
| Returns true if the graph is weighted (valued), i.e. if any e in |E| has value not 0 or 1 Complexity: O(n^2) | |
| void | setWeighted (const bool &toggle=true) |
| Sets the graph to be weighted ( valued edges ). | |
| qreal | graphReciprocity () |
| Gets the arc reciprocity of the graph. | |
| bool | isSymmetric () |
| Returns TRUE if the adjacency matrix of the current relation is symmetric. | |
| void | setSymmetric () |
| Transforms the graph to symmetric (all edges reciprocal) | |
| void | addRelationSymmetricStrongTies (const bool &allRelations=false) |
| Creates a new symmetric relation by keeping only strong-ties (mutual links) in the current relation. In the new relation, two actors are connected only if they are mutually connected in the current relation. | |
| void | relationAddCocitation () |
| Creates a new symmetric relation by connecting those actors that are cocitated by others. In the new relation, an edge will exist between actor i and actor j only if C(i,j) > 0, where C the Cocitation Matrix. Thus the actor pairs cited by more common neighbors will appear with a stronger tie between them than pairs those cited by fewer common neighbors. The resulting relation is symmetric. | |
| void | graphDichotomization (const qreal threshold) |
| Creates a new binary relation in a valued network using edge dichotomization according to the threshold parameter. | |
| void | setDirected (const bool &toggle=true, const bool &signalMW=true) |
| Toggles the graph directed or undirected. | |
| void | setUndirected (const bool &toggle=true, const bool &signalMW=true) |
| Makes the graph undirected or directed. | |
| bool | isDirected () |
| Returns true if graph is directed. | |
| bool | isUndirected () |
| Returns true if graph is undirected. | |
| bool | isConnected () |
| Checks if the graph is connected, in the sense of a topological space, i.e., there is a path from any vertex to any other vertex in the graph. Called from MW::slotConnectedness() | |
| void | createMatrixAdjacency (const bool dropIsolates=false, const bool considerWeights=true, const bool inverseWeights=false, const bool symmetrize=false) |
| Creates an adjacency matrix AM where AM(i,j)=1 if i is connected to j and AM(i,j)=0 if i not connected to j Used in Graph::centralityInformation(), Graph::graphWalksMatrixCreate and Graph::createMatrixAdjacencyInverse() | |
| bool | createMatrixAdjacencyInverse (const QString &method="lu") |
| Computes the inverse of the current adjacency matrix. | |
| void | createMatrixSimilarityMatching (Matrix &AM, Matrix &SEM, const int &measure=METRIC_SIMPLE_MATCHING, const QString &varLocation="Rows", const bool &diagonal=false, const bool &considerWeights=true) |
| Calls Matrix:similarityMatrix to compute the similarity matrix SCM of the variables (rows, columns, both) in given input matrix using the selected matching measure. | |
| void | createMatrixSimilarityPearson (Matrix &AM, Matrix &PCC, const QString &varLocation="Rows", const bool &diagonal=false) |
| The Pearson product-moment correlation coefficient (PPMCC, PCC or Pearson's r) is a measure of the linear dependence between two variables X and Y. | |
| void | createMatrixDissimilarities (Matrix &INPUT_MATRIX, Matrix &DSM, const int &metric, const QString &varLocation, const bool &diagonal, const bool &considerWeights) |
| Calls Matrix:distancesMatrix to compute the dissimilarities matrix DSM of the variables (rows, columns, both) in given input matrix using the user defined metric. | |
| void | setReportsDataDir (const QString &reportsDir) |
| Sets the directory where reports are saved This is used when exporting prominence distribution images to be used in HTML reports. | |
| void | setReportsRealNumberPrecision (const int &precision) |
| Sets the precision (number of fraction digits) the app will use when writing real numbers in reports. | |
| void | setReportsLabelLength (const int &length) |
| Sets the length of labels in reports. | |
| void | setReportsChartType (const int &type) |
| Sets the chart type in reports. | |
| void | writeDataSetToFile (const QString dir, const QString) |
| Writes a "famous" dataset to the given file Datasets are hardcoded! They are exported in the given fileName... | |
| void | writeMatrixAdjacencyTo (QTextStream &os, const bool &saveEdgeWeights=true) |
| void | writeReciprocity (const QString fileName, const bool considerWeights=false) |
| Writes reciprocity report to filename. | |
| void | writeMatrix (const QString &fileName, const int &matrix=MATRIX_ADJACENCY, const bool &considerWeights=true, const bool &inverseWeights=false, const bool &dropIsolates=false, const QString &varLocation="Rows", const bool &simpler=false) |
| void | writeMatrixHTMLTable (QTextStream &outText, Matrix &M, const bool &markDiag=true, const bool &plain=false, const bool &printInfinity=true, const bool &dropIsolates=false) |
| Writes the matrix M as HTML. | |
| void | writeMatrixAdjacency (const QString fileName, const bool &markDiag=true) |
| void | writeMatrixAdjacencyPlot (const QString fileName, const bool &simpler=false) |
| Writes a visual representation of the adjacency matrix of the graph to the specified file. | |
| void | writeMatrixAdjacencyInvert (const QString &filename, const QString &method) |
| Calls Graph::createMatrixAdjacencyInverse(method) to compute the inverse of the adjacency matrix and writes it to file fn in HTML notation. | |
| void | writeMatrixLaplacianPlainText (const QString &filename) |
| Computes the Laplacian matrix of the graph and writes it to given file. | |
| void | writeMatrixDegreeText (const QString &filename) |
| Computes the Degree matrix of the graph and writes it to given file. | |
| void | writeMatrixDistancesPlainText (const QString &fn, const bool &considerWeights, const bool &inverseWeights, const bool &dropIsolates) |
| Writes the matrix of distances to a file. | |
| void | writeMatrixShortestPathsPlainText (const QString &fn, const bool &considerWeights, const bool &inverseWeights) |
| Writes the shortest paths matrix to a file Each SIGMA(i,j) element is the number of shortest paths (geodesics) from i and j. | |
| void | writeMatrixDissimilarities (const QString fileName, const QString &metricStr, const QString &varLocation, const bool &diagonal, const bool &considerWeights) |
| Writes dissimilarity matrix based on a metric/measure to given html file. | |
| void | writeMatrixSimilarityMatchingPlain (const QString fileName, const int &measure=METRIC_SIMPLE_MATCHING, const QString &matrix="adjacency", const QString &varLocation="rows", const bool &diagonal=false, const bool &considerWeights=true) |
| Writes similarity matrix based on a matching measure to given file. | |
| void | writeMatrixSimilarityMatching (const QString fileName, const QString &measure="Simple", const QString &matrix="adjacency", const QString &varLocation="rows", const bool &diagonal=false, const bool &considerWeights=true) |
| Writes similarity matrix based on a matching measure to given html file. | |
| void | writeMatrixSimilarityPearson (const QString fileName, const bool considerWeights, const QString &matrix="adjacency", const QString &varLocation="rows", const bool &diagonal=false) |
| Calls Graph::createMatrixSimilarityPearson() and writes Pearson Correlation Coefficients to given file. | |
| void | writeMatrixSimilarityPearsonPlainText (const QString fileName, const bool considerWeights, const QString &matrix="adjacency", const QString &varLocation="rows", const bool &diagonal=false) |
| Calls Graph::graphSimilariyPearsonCorrelationCoefficients() and writes Pearson Correlation Coefficients to given file. | |
| void | writeEccentricity (const QString fileName, const bool considerWeights=false, const bool inverseWeights=false, const bool dropIsolates=false) |
| Writes the Eccentricity report to file. | |
| void | writeCentralityDegree (const QString, const bool weights, const bool dropIsolates) |
| Writes the Degree Centrality to a file. | |
| void | writeCentralityCloseness (const QString, const bool weights, const bool inverseWeights, const bool dropIsolates) |
| Writes the closeness centralities to a file. | |
| void | writeCentralityClosenessInfluenceRange (const QString, const bool weights, const bool inverseWeights, const bool dropIsolates) |
| Writes the "improved" closeness centrality indices to a file. | |
| void | writeCentralityBetweenness (const QString, const bool weights, const bool inverseWeights, const bool dropIsolates) |
| Writes Betweenness centralities to file. | |
| void | writeCentralityPower (const QString, const bool weigths, const bool inverseWeights, const bool dropIsolates) |
| Writes Power Centralities to a file. | |
| void | writeCentralityStress (const QString, const bool weigths, const bool inverseWeights, const bool dropIsolates) |
| Writes the Stress centralities to a file. | |
| void | writeCentralityEccentricity (const QString, const bool weigths, const bool inverseWeights, const bool dropIsolates) |
| Writes the Eccentricity centralities (aka Harary Graph Centrality) to a file. | |
| void | writeCentralityInformation (const QString, const bool weigths, const bool inverseWeights) |
| Writes the information centralities to file. | |
| void | writeCentralityEigenvector (const QString, const bool &weigths=true, const bool &inverseWeights=false, const bool &dropIsolates=false) |
| Writes the eigenvector centralities to a file. | |
| void | writePrestigeDegree (const QString, const bool weights, const bool dropIsolates) |
| Writes the Degree Prestige of each node to a file. | |
| void | writePrestigeProximity (const QString, const bool weights, const bool inverseWeights, const bool dropIsolates) |
| Writes the proximity prestige indices to a file. | |
| void | writePrestigePageRank (const QString, const bool Isolates=false) |
| Writes the PageRank scores of vertices to a file. | |
| bool | writeClusteringHierarchical (const QString &fileName, const QString &varLocation, const QString &matrix="Adjacency", const QString &metric="Manhattan", const QString &method="Complete", const bool &diagonal=false, const bool &dendrogram=false, const bool &considerWeights=true, const bool &inverseWeights=false, const bool &dropIsolates=false) |
| Writes Hierarchical Clustering Analysis to a given file. | |
| void | writeClusteringHierarchicalResultsToStream (QTextStream &outText, const int N, const bool &dendrogram=false) |
| Writes Hierarchical Clustering results to given output stream Before running this methos, the method Graph::graphClusteringHierarchical() must execute and return true. Otherwise, the result is unpredictable... | |
| bool | writeCliqueCensus (const QString &fileName, const bool considerWeights) |
| Graph::writeCliqueCensus Calls graphCliques() to compute all cliques (maximal connected subgraphs) of the network. Then writes the results into a file, along with the Actor by clique analysis, the Co-membership matrix and the Hierarchical clustering of overlap matrix. | |
| void | writeClusteringCoefficient (const QString, const bool) |
| Graph::writeClusteringCoefficient Writes the clustering coefficients to a file. | |
| void | writeTriadCensus (const QString, const bool) |
| int | graphConnectednessFull (const bool updateProgress=false) |
| bool | graphReachable (const int &v1, const int &v2) |
| Returns true if vertices v1 and v2 are reachable. | |
| void | createMatrixReachability () |
| Creates the reachability matrix XRM. | |
| int | graphDiameter (const bool considerWeights, const bool inverseWeights) |
| Returns the diameter of the graph, aka the largest geodesic distance between any two vertices. | |
| int | graphDistanceGeodesic (const int &v1, const int &v2, const bool &considerWeights=false, const bool &inverseWeights=true) |
| Returns the geodesic distance (length of shortest path) from vertex v1 to vertex v2. | |
| qreal | graphDistanceGeodesicAverage (const bool considerWeights, const bool inverseWeights, const bool dropIsolates) |
| Returns the average distance of the graph. | |
| void | graphDistancesGeodesic (const bool &computeCentralities=false, const bool &considerWeights=false, const bool &inverseWeights=true, const bool &dropIsolates=false) |
| Computes the geodesic distances between all vertices: In the process, it also computes many other centrality/prestige metrics: | |
| void | graphMatrixDistanceGeodesicCreate (const bool &considerWeights=false, const bool &inverseWeights=true, const bool &dropIsolates=false) |
| Creates the matrix DM of geodesic distances between vertices. | |
| void | graphMatrixShortestPathsCreate (const bool &considerWeights=false, const bool &inverseWeights=true, const bool &dropIsolates=false) |
| Creates the matrix SIGMA of shortest paths (geodesics) between vertices Each SIGMA(i,j) is the number of shortest paths (geodesics) from i and j. | |
| int | getProminenceIndexByName (const QString &prominenceIndexName) |
| Returns the IndexType of the given prominence index name Called from MW::slotEditNodeFind, MW::slotLayoutRadialByProminenceIndex etc. | |
| void | prominenceDistribution (const int &index, const ChartType &type, const QString &distImageFileName=QString()) |
| Computes the distribution of a centrality index score. The distribution is stored as Qt Series depending on the SeriesType parameter type It is send to MW through signal/slot. | |
| void | prominenceDistributionBars (const H_StrToInt &discreteClasses, const QString &name, const QString &distImageFileName) |
| Computes the distribution of a centrality index scores. The distribution data are returned as QBarSeries series (with a QBarSet attached) to MW which in turn displays them on a Bar Chart. | |
| void | prominenceDistributionArea (const H_StrToInt &discreteClasses, const QString &name, const QString &distImageFileName) |
| Computes the distribution of a centrality index scores. The distribution data are returned as QAreaSeries series to MW which in turn displays them on a Area Chart. | |
| void | prominenceDistributionSpline (const H_StrToInt &discreteClasses, const QString &seriesName, const QString &distImageFileName) |
| Computes the distribution of a centrality index scores. The distribution data are returned as QSplineSeries series to MW which in turn displays them on a Spline Chart. | |
| void | centralityDegree (const bool &considerWeights=true, const bool &dropIsolates=false) |
| Calculates the degree (outDegree) centrality of each vertex - diagonal included. | |
| void | centralityInformation (const bool considerWeights=false, const bool inverseWeights=false) |
| Computes the Information centrality of each vertex - diagonal included Note that there is no known generalization of Stephenson&Zelen's theory for information centrality to directional data. | |
| void | centralityEigenvector (const bool &considerWeights=false, const bool &inverseWeights=false, const bool &dropIsolates=false) |
| Computes Eigenvector centrality. | |
| void | centralityClosenessIR (const bool considerWeights=false, const bool inverseWeights=false, const bool dropIsolates=false) |
| Computes an "improved" closeness centrality index, IRCC, which can be used on disconnected graphs. IRCC is an improved node-level centrality closeness index which focuses on the influence range of each node (the set of nodes that are reachable from it) For each node v, this index calculates the fraction of nodes in its influence range and divides it by the average distance of those nodes from v, ignoring nodes that are not reachable from it. | |
| void | prestigeDegree (const bool &considerWeights, const bool &dropIsolates=false) |
| Computes the Degree Prestige (in-degree) of each vertex - diagonal included Also the mean value and the variance of the in-degrees. | |
| void | prestigePageRank (const bool &dropIsolates=false) |
| Calculates the PageRank Prestige of each vertex. | |
| void | prestigeProximity (const bool considerWeights=false, const bool inverseWeights=false, const bool dropIsolates=false) |
| Computes Proximity Prestige of each vertex Also the mean value and the variance of it.. | |
| int | walksBetween (int v1, int v2, int length) |
| Calculates and returns the number of walks of a given length between v1 and v2. | |
| void | graphWalksMatrixCreate (const int &N=0, const int &length=0, const bool &updateProgress=false) |
| Computes either the "Walks of given length" or the "Total Walks" matrix. If length>0, it computes the Walks of given length matrix, XM=AM^l where each element (i,j) denotes the number of walks of length l between vertex i and j. If length=0, it computes the Total Walks matrix, XSM=Sum{AM^n} where each (i,j) denotes the total number of walks of any length between vertices i and j. NOTE: In the latter case, this function is VERY SLOW on large networks (n>50), since it will calculate all powers of the sociomatrix up to n-1 in order to find out all possible walks. | |
| void | writeWalksTotalMatrixPlainText (const QString &fn) |
| Graph::writeWalksTotalMatrixPlainText Writes the total number of walks matrix. | |
| void | writeWalksOfLengthMatrixPlainText (const QString &fn, const int &length) |
| Graph::writeWalksOfLengthMatrixPlainText. | |
| void | writeMatrixWalks (const QString &fn, const int &length=0, const bool &simpler=false) |
| Calls graphWalksMatrixCreate() to compute walks and writes the Walks of given length matrix to a file in HTML. If length = 0, it writes the Total Walks matrix. | |
| QList< int > | vertexinfluenceRange (int v1) |
| Returns the influence range of vertex v1, namely the set of nodes who are reachable from v1 (See Wasserman and Faust, pp.200-201, based on Lin, 1976). The Influence Range of vertex v can also be defined as: Ji = Sum [ D(v,j), iff D(v,j) != inf ] for every j in V, where j!=v and D the distance matrix This function is for digraphs only. | |
| QList< int > | vertexinfluenceDomain (int v2) |
| Returns the influence domain of vertex v1, namely the set of nodes who can reach v1 The Influence Domain Ii of vertex v can also be defined as: Ii = Sum [ D(i,v), iff D(i,v) != inf ] for every in V, where i!=v and D the distance matrix This function applies to digraphs only. | |
| void | writeReachabilityMatrixPlainText (const QString &fn, const bool &dropIsolates=false) |
| qreal | numberOfTriples (int v1) |
| void | graphCliques (QSet< int > R=QSet< int >(), QSet< int > P=QSet< int >(), QSet< int > X=QSet< int >()) |
| Finds all maximal cliques in an undirected (?) graph. Implements the Bron–Kerbosch algorithm, a recursive backtracking algorithm that searches for all maximal cliques in a given graph G. Given three sets R, P, and X, the algorithm finds the maximal cliques that include all of the vertices in R, some of the vertices in P, and none of the vertices in X. In each call to the algorithm, P and X are disjoint sets whose union consists of those vertices that form cliques when added to R. In other words, P ∪ X is the set of vertices which are joined to every element of R. When P and X are both empty there are no further elements that can be added to R, so R is a maximal clique and the algorithm outputs R. The recursion is initiated by setting R and X to be the empty set and P to be the vertex set of the graph. Within each recursive call, the algorithm considers the vertices in P in turn. if there are no vertices, it either reports R as a maximal clique (if X is empty), or backtracks. For each vertex v chosen from P, it makes a recursive call in which v is added to R and in which P and X are restricted to the neighbor set NBS(v) of v, which finds and reports all clique extensions of R that contain v. Then, it moves v from P to X to exclude it from consideration in future cliques and continues with the next vertex in P. | |
| void | graphCliqueAdd (const QList< int > &clique) |
| Called from Graph::graphCliques to add a new clique (list of vertices) Adds clique info to each clique member and updates co-membership matrix CLQM . | |
| int | graphCliquesContaining (const int &actor, const int &size=0) |
| int | graphCliquesOfSize (const int &size) |
| Graph::graphCliquesOfSize Returns the number of maximal cliques of a given size. | |
| bool | graphClusteringHierarchical (Matrix &STR_EQUIV, const QString &varLocation, const int &metric, const int &method, const bool &diagonal=false, const bool &diagram=false, const bool &considerWeights=true, const bool &inverseWeights=false, const bool &dropIsolates=false) |
| Performs an hierarchical clustering process (Johnson, 1967) on a given NxN distance/dissimilarity matrix. The input matrix can be the the adjacency matrix, the geodesic distance matrix or a derived from them dissimilarities matrix using a user-specified metric, i.e. euclidean distance. The method parameter defines how to compute distances (similarities) between a new cluster the old clusters. Valid values can be: | |
| qreal | clusteringCoefficientLocal (const int &v1) |
| Returns the local clustering coefficient (CLUCOF) of a vertex v1. | |
| qreal | clusteringCoefficient (const bool updateProgress=false) |
| Computes local clustering coefficients and returns the network average Clustering Coefficient. | |
| bool | graphTriadCensus () |
| Graph::graphTriadCensus Conducts a triad census and updates QList::triadTypeFreqs, which is the list carrying all triad type frequencies Complexity:O(n!) | |
| void | triadType_examine_MAN_label (int, int, int, GraphVertex *, GraphVertex *, GraphVertex *) |
| void | layoutRandom () |
| Repositions all nodes on random positions Emits setNodePos(i, x,y) to tell GW that the node item should be moved. | |
| void | layoutRadialRandom (const bool &guides=true) |
| Repositions all nodes on the periphery of different circles with random radius. | |
| void | layoutCircular (const double &x0, const double &y0, const double &newRadius, const bool &guides=false) |
| Repositions all nodes on the periphery of a circle with given radius. | |
| void | layoutByProminenceIndex (int prominenceIndex, int layoutType, const bool &considerWeights=false, const bool &inverseWeights=false, const bool &dropIsolates=false) |
| Applies a layout according to each actor's prominence index score. The layout type can be radial (0), level (1), node sizes (2) or node colors (3), as follows: layoutType=0 - Repositions all nodes on the periphery of concentric circles with radius analogous to their prominence index layoutType=1 - Repositions all nodes on different top-down levels according to their centrality layoutType=2 - Changes node sizes to be proportional to their prominence index score layoutType=2 - Changes node colors to reflect their prominence index score (from red to green) | |
| void | layoutVertexSizeByIndegree () |
| Convenience method Changes the size of all nodes to be proportional to their InDegree (Degree Prestige) Calls layoutByProminenceIndex. | |
| void | layoutVertexSizeByOutdegree () |
| Convenience method Changes the size of all nodes to be proportional to their outDegree (Degree Centrality) Calls layoutByProminenceIndex. | |
| void | layoutForceDirectedSpringEmbedder (const int maxIterations) |
| Embeds a Force Directed Placement layout according to the initial Spring Embedder model proposed by Eades. | |
| void | layoutForceDirectedFruchtermanReingold (const int maxIterations) |
| Embeds a Force Directed Placement layout according to the Fruchterman-Reingold model. Fruchterman and Reingold (1991) refined the Spring Embedder model by replacing the forces. In this model, "the vertices behave as atomic particles or celestial bodies,
exerting attractive and repulsive forces on one another." (ibid). Again, only vertices that are neighbours attract each other but, unlike Spring Embedder, all vertices repel each other. These forces induce movement. The algorithm might resemble molecular or planetary simulations, sometimes called n-body problems. | |
| void | layoutForceDirectedKamadaKawai (const int maxIterations=500, const bool considerWeights=false, const bool inverseWeights=false, const bool dropIsolates=false, const QString &initialPositions="current") |
| Embeds a Force Directed Placement layout according to the Kamada-Kawai model. In this model, the network is considered to be a dynamic system where every two actors are 'particles' mutually connected by a 'spring'. Each spring has a desirable length, which corresponds to their graph theoretic distance. In this way, the optimal layout of the graph is the state with the minimum imbalance. The degree of imbalance is formulated as the total spring energy: the square summation of the differences between desirable distances and real ones for all pairs of particles Initially, the particles/actors are placed on the vertices of a regular n-polygon. | |
| qreal | graphDistanceEuclidean (const QPointF &a, const QPointF &b) |
| Computes the euclideian distance between QPointF a and b. | |
| qreal | graphDistanceEuclidean (const QPointF &a) |
| the euclideian distance of QPointF a (where a is difference vector) | |
| int | sign (const qreal &D) |
| Graph::sign returns the sign of number D as integer (1 or -1) | |
| qreal | layoutForceDirected_F_rep (const QString model, const qreal &dist, const qreal &optimalDistance) |
| qreal | layoutForceDirected_F_att (const QString model, const qreal &dist, const qreal &optimalDistance) |
| void | layoutForceDirected_Eades_moveNodes (const qreal &c4) |
| void | layoutForceDirected_FR_moveNodes (const qreal &temperature) |
| Graph::layoutForceDirected_FR_moveNodes. | |
| qreal | layoutForceDirected_FR_temperature (const int iteration) const |
| Reduces the temperature as the layout approaches a better configuration. | |
| qreal | computeOptimalDistance (const int &V) |
| Computes Optimal Distance. Used in Spring Embedder and FR algorithms. | |
| void | compute_angles (const QPointF &Delta, const qreal &dist, qreal &angle1, qreal &angle2, qreal °rees1, qreal °rees2) |
| Graph::compute_angles Computes the two angles of the orthogonal triangle shaped by two points of difference vector DV and distance dist A = 90 B = angle1 C = angle2. | |
| void | webCrawlTerminateThreads (QString reason) |
| called from Graph, when closing network, to terminate all crawler processes Also called indirectly when wc_spider finishes | |
| void | randomizeThings () |
| Adds a little universal randomness :) | |
| void | randomNetErdosCreate (const int &N, const QString &model, const int &m, const qreal &p, const QString &mode, const bool &diag) |
| Creates an erdos-renyi random network according to the given model. | |
| void | randomNetScaleFreeCreate (const int &N, const int &power, const int &m0, const int &m, const qreal &alpha, const QString &mode) |
| Creates a scale-free random-network network. | |
| void | randomNetSmallWorldCreate (const int &N, const int °ree, const double &beta, const QString &mode) |
| Creates a small world random network. | |
| void | randomNetRingLatticeCreate (const int &N, const int °ree, const bool updateProgress=false) |
| Creates a random ring lattice network. | |
| void | randomNetRegularCreate (const int &N, const int °ree, const QString &mode, const bool &diag) |
| Creates a random network where nodes have the same degree. | |
| void | randomNetLatticeCreate (const int &N, const int &length, const int &dimension, const int &neighborhoodLength, const QString &mode, const bool &circular=false) |
| Creates a lattice network. | |
| int | factorial (int) |
Data Fields | |
| H_Int | vpos |
| H_f_i | sizeOfNthOrderNeighborhood |
Private Member Functions | |
| void | edgeAdd (const int &v1, const int &v2, const qreal &weight, const int &type, const QString &label, const QString &color) |
| Adds a directed edge from v1 to v2 If type == EdgeType::Undirected then it also adds the directed edge v2->v1. | |
| void | BFS (const int &s, const int &si, const bool &computeCentralities=false, const bool &dropIsolates=false) |
| void | dijkstra (const int &s, const int &si, const bool &computeCentralities=false, const bool &inverseWeights=false, const bool &dropIsolates=false) |
| void | minmax (qreal C, GraphVertex *v, qreal &max, qreal &min, int &maxNode, int &minNode) |
| Computes minimum and maximum centralities during graphDistancesGeodesic() | |
| void | resolveClasses (qreal C, H_StrToInt &discreteClasses, int &classes) |
| Checks if score C is a new prominence class If yes, it stores that number in a QHash<QString,int> type where the score is the key. If no, increases the frequency of this prominence score by 1 Called from graphDistancesGeodesic() | |
| void | resolveClasses (qreal C, H_StrToInt &discreteClasses, int &classes, int name) |
| Overloaded method. It only adds displaying current vertex for debugging purposes. | |
The Graph class This is the main class for a Graph, used in conjuction with GraphVertex, Parser and Matrix objects. Graph class methods are the interface to various analysis algorithms GraphVertex class holds each vertex data (colors, strings, statistics, etc) Matrix class holds the adjacency matrix of the network. Parser class loads files of networks.
KNOWN BUGS:
wontfix Cannot read Graphml files where nodes are not declared before edges (i.e. nets/killer.graphml)
wontfix Pajek files with no ic / labels without quotes are displayed without colors
todo Group edge editing: i.e. change weight or color. todo Subgroups / Communities: clans, path distance MDS, Components, Blocks and Cutpoints. todo Structural Equivalence: MDS, Block modelling, CONCOR
| enum Graph::Clustering |
| enum Graph::ModStatus |
| Graph::Graph | ( | const int & | reserveVerticesSize = 5000, |
| const int & | reserveEdgesPerVertexSize = 500 |
||
| ) |
Constructs a Graph.
| Graph::~Graph | ( | ) |
Destroys the Graph object.
|
signal |
|
signal |
| void Graph::addRelationSymmetricStrongTies | ( | const bool & | allRelations = false | ) |
Creates a new symmetric relation by keeping only strong-ties (mutual links) in the current relation. In the new relation, two actors are connected only if they are mutually connected in the current relation.
| allRelations |
|
private |
methods used by graphDistancesGeodesic()
Breadth-First Search (BFS) method for unweighted graphs (directed or not)
INPUT: a 'source' vertex with vpos s and a boolean computeCentralities. (Implicitly, BFS uses the m_graph structure)
OUTPUT: For every vertex t: d(s, t) is set to the distance of each t from s For every vertex t: s(s, t) is set to the number of shortest paths between s and t
Also, if computeCentralities is true then BFS does extra operations: a) For source vertex s: it calculates CC(s) as the sum of its distances from every other vertex. it calculates eccentricity(s) as the maximum distance from all other vertices. it increases sizeOfNthOrderNeighborhood [ N ] by one, to store the number of nodes at distance n from source s b) For every vertex u: it increases SC(u) by one, when it finds a new shor. path from s to t through u. appends each neighbor y of u to the list , thus Ps stores all predecessors of y on all all shortest paths from s c) Each vertex u popped from Q is pushed to a stack Stack
|
slot |
Gets the max radius of the canvas.
|
slot |
Gets the min dimensions of the canvas.
|
slot |
Returns a random x-coordinate adjusted to be visible inside the canvas usable area.
|
slot |
Returns a random y-coordinate adjusted to be visible inside the canvas usable area.
|
slot |
Sets the size of the canvas.
Called when the MW is resized to update canvasWidth/canvasHeight, and node positions
| w | |
| h |
|
slot |
Checks if x is visible inside the canvas usable area and if not returns an adjusted x-coordinate.
| x |
|
slot |
Checks if y is visible inside the canvas usable area and if not returns an adjusted y-coordinate.
| y |
| void Graph::centralityClosenessIR | ( | const bool | considerWeights = false, |
| const bool | inverseWeights = false, |
||
| const bool | dropIsolates = false |
||
| ) |
Computes an "improved" closeness centrality index, IRCC, which can be used on disconnected graphs. IRCC is an improved node-level centrality closeness index which focuses on the influence range of each node (the set of nodes that are reachable from it) For each node v, this index calculates the fraction of nodes in its influence range and divides it by the average distance of those nodes from v, ignoring nodes that are not reachable from it.
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::centralityDegree | ( | const bool & | considerWeights = true, |
| const bool & | dropIsolates = false |
||
| ) |
Calculates the degree (outDegree) centrality of each vertex - diagonal included.
| considerWeights | |
| dropIsolates |
| void Graph::centralityEigenvector | ( | const bool & | considerWeights = false, |
| const bool & | inverseWeights = false, |
||
| const bool & | dropIsolates = false |
||
| ) |
Computes Eigenvector centrality.
| considerWeights | |
| inverseWeights |
| void Graph::centralityInformation | ( | const bool | considerWeights = false, |
| const bool | inverseWeights = false |
||
| ) |
Computes the Information centrality of each vertex - diagonal included Note that there is no known generalization of Stephenson&Zelen's theory for information centrality to directional data.
| considerWeights | |
| inverseWeights |
| void Graph::clear | ( | const QString & | reason = "" | ) |
Clears all vertices.
| reason |
| qreal Graph::clusteringCoefficient | ( | const bool | updateProgress = false | ) |
Computes local clustering coefficients and returns the network average Clustering Coefficient.
| updateProgress |
| qreal Graph::clusteringCoefficientLocal | ( | const int & | v1 | ) |
Returns the local clustering coefficient (CLUCOF) of a vertex v1.
| v1 |
| void Graph::compute_angles | ( | const QPointF & | DV, |
| const qreal & | dist, | ||
| qreal & | angle1, | ||
| qreal & | angle2, | ||
| qreal & | degrees1, | ||
| qreal & | degrees2 | ||
| ) |
Graph::compute_angles Computes the two angles of the orthogonal triangle shaped by two points of difference vector DV and distance dist A = 90 B = angle1 C = angle2.
| DV | |
| dist | |
| angle1 | |
| angle2 | |
| degrees1 | |
| degrees2 |
| qreal Graph::computeOptimalDistance | ( | const int & | V | ) |
Computes Optimal Distance. Used in Spring Embedder and FR algorithms.
| void Graph::createMatrixAdjacency | ( | const bool | dropIsolates = false, |
| const bool | considerWeights = true, |
||
| const bool | inverseWeights = false, |
||
| const bool | symmetrize = false |
||
| ) |
Creates an adjacency matrix AM where AM(i,j)=1 if i is connected to j and AM(i,j)=0 if i not connected to j Used in Graph::centralityInformation(), Graph::graphWalksMatrixCreate and Graph::createMatrixAdjacencyInverse()
| dropIsolates | |
| considerWeights | |
| inverseWeights | |
| symmetrize |
| bool Graph::createMatrixAdjacencyInverse | ( | const QString & | method = "lu" | ) |
Computes the inverse of the current adjacency matrix.
| method |
| void Graph::createMatrixDissimilarities | ( | Matrix & | INPUT_MATRIX, |
| Matrix & | DSM, | ||
| const int & | metric, | ||
| const QString & | varLocation, | ||
| const bool & | diagonal, | ||
| const bool & | considerWeights | ||
| ) |
Calls Matrix:distancesMatrix to compute the dissimilarities matrix DSM of the variables (rows, columns, both) in given input matrix using the user defined metric.
| INPUT_MATRIX | |
| DSM | |
| metric | |
| varLocation | |
| diagonal | |
| considerWeights |
| void Graph::createMatrixReachability | ( | ) |
Creates the reachability matrix XRM.
| void Graph::createMatrixSimilarityMatching | ( | Matrix & | AM, |
| Matrix & | SCM, | ||
| const int & | measure = METRIC_SIMPLE_MATCHING, |
||
| const QString & | varLocation = "Rows", |
||
| const bool & | diagonal = false, |
||
| const bool & | considerWeights = true |
||
| ) |
Calls Matrix:similarityMatrix to compute the similarity matrix SCM of the variables (rows, columns, both) in given input matrix using the selected matching measure.
| AM | |
| SCM | |
| rows |
| void Graph::createMatrixSimilarityPearson | ( | Matrix & | AM, |
| Matrix & | PCC, | ||
| const QString & | varLocation = "Rows", |
||
| const bool & | diagonal = false |
||
| ) |
The Pearson product-moment correlation coefficient (PPMCC, PCC or Pearson's r) is a measure of the linear dependence between two variables X and Y.
As a normalized version of the covariance, the PPMCC is computed with the formula: r =\frac{\sum ^n _{i=1}(x_i - \bar{x})(y_i - \bar{y})}{\sqrt{\sum ^n _{i=1}(x_i - \bar{x})^2} \sqrt{\sum ^n _{i=1}(y_i - \bar{y})^2}}
It gives a value between +1 and −1 inclusive, where 1 is total positive linear correlation, 0 is no linear correlation, and −1 is total negative linear correlation.
In SNA, Pearson correlations can be used to track the similarity between actors, in terms of structural equivalence.
This method creates an actor by actor NxN matrix PCC where the (i,j) element is the Pearson correlation coefficient of actor i and actor j. If the input matrix is the adjacency matrix, the PCC of two nodes measures how related (similar, inverse or not related at all) their patterns of ties tend to be. A positive value means there is strong linear association of the two actors, while a negative value means the inverse. For instance a value of -1 means the two actors have exactly opposite ties to other actors, while a value of 1 means the actors have identical patterns of ties to other actors (they are connected to the same actors).
The correlation measure of similarity is particularly useful when the data on ties are valued
| AM | |
| PCC | |
| rows |
|
private |
Dijkstra's algorithm for solving the SSSP problem in weighted graphs (directed or not). It uses a min-priority queue prQ to provide constant time lookup of the minimum distance. The priority queue is implemented with std::priority_queue
INPUT: a 'source' vertex with vpos s and a boolean computeCentralities. (Implicitly, the algorithm uses the m_graph structure)
OUTPUT: For every vertex t: d(s, t) is set to the distance of each t from s For every vertex t: s(s, t) is set to the number of shortest paths between s and t
Also, if computeCentralities is true then it does extra operations: a) For source vertex s: it calculates CC(s) as the sum of its distances from every other vertex. it calculates eccentricity(s) as the maximum distance from all other vertices. it increases sizeOfNthOrderNeighborhood [ N ] by one, to store the number of nodes at distance n from source s b) For every vertex u: it increases SC(u) by one, when it finds a new shor. path from s to t through u. appends each neighbor y of u to the list Ps, thus Ps stores all predecessors of y on all all shortest paths from s c) Each vertex u popped from prQ is pushed to a stack Stack
|
private |
Adds a directed edge from v1 to v2 If type == EdgeType::Undirected then it also adds the directed edge v2->v1.
private member functions
| v1 | |
| v2 | |
| weight | |
| label | |
| color | |
| type |
| MyEdge Graph::edgeClicked | ( | ) |
Returns clicked edge.
|
slot |
Sets the clicked edge.
Parameters are the source and target node of the edge. It emits signal to MW, which displays a relevant message on the status bar.
| v1 | |
| v2 |
| QString Graph::edgeColor | ( | const int & | v1, |
| const int & | v2 | ||
| ) |
Returns the color of the directed edge v1->v2.
| v1 | |
| v2 |
| bool Graph::edgeColorAllSet | ( | const QString & | color, |
| const int & | threshold = RAND_MAX |
||
| ) |
Changes the color of all enabled edges.
| color |
| void Graph::edgeColorInit | ( | const QString & | color | ) |
Saves the default edge color Used by random network creation methods.
| color |
| void Graph::edgeColorSet | ( | const int & | v1, |
| const int & | v2, | ||
| const QString & | color | ||
| ) |
Changes the color of edge v1->v2.
| v1 | |
| v2 | |
| color |
|
slot |
Checks a) if edge exists and b) if the reverse edge exists Calls edgeAdd to add the new edge to the Graph, then emits drawEdge() which calls GW::drawEdge() to draw the new edge. Called from homonymous signal of Parser class. Also called from MW when user clicks on the "add link" button Also called (via MW) from GW when user middle-clicks on two nodes.
Slots to signals from GraphicsWidget and Parser
| v1 | |
| v2 | |
| weight | |
| color | |
| reciprocal | |
| drawArrows | |
| bezier |
|
slot |
Called from WebCrawler when it finds an new link Calls edgeCreate() method with initEdgeColor.
| source | |
| target |
| qreal Graph::edgeExists | ( | const int & | v1, |
| const int & | v2, | ||
| const bool & | checkReciprocal = false |
||
| ) |
Checks if there is an edge from v1 to v2 and returns the weight, if the edge exists.
Complexity: O(logN) for vpos retrieval + O(1) for QList index retrieval + O(logN) for checking edge(v2)
| v1 | |
| v2 | |
| reciprocated | if true, checks if the edge is reciprocated (v1<->v2) with the same weight |
| qreal Graph::edgeExistsVirtual | ( | const int & | v1, |
| const int & | v2 | ||
| ) |
Checks if there is an edge from v1 to v2, even weight = 0 and returns the weight, if the edge exists or RAND_MAX if the edge does not exist at all.
This is only used in GraphML saving if the user has selected the Settings option to save zero-weight edges
| v1 | |
| v2 |
|
slot |
Toggles (enables or disables) all edges of the given relation.
Calls the homonymous method of GraphVertex class.
| relation | |
| status |
|
slot |
Filters (disables) edges according the specified threshold weight.
| m_threshold | |
| overThreshold |
|
slot |
Enables or disables unilateral edges in current relationship.
If toggle=true, all non-reciprocal edges are disabled, effectively making the network symmetric.
| toggle |
| void Graph::edgeInboundStatusSet | ( | const int & | target, |
| const int & | source, | ||
| const bool & | toggle = false |
||
| ) |
Toggles the status of inbound edge target <- source at target vertex.
| v1 | |
| v2 | |
| toggle |
| QString Graph::edgeLabel | ( | const int & | v1, |
| const int & | v2 | ||
| ) | const |
Returns the label of edge v1->v2.
| v1 | |
| v2 |
| void Graph::edgeLabelSet | ( | const int & | v1, |
| const int & | v2, | ||
| const QString & | label | ||
| ) |
Changes the label of edge v1->v2.
| v1 | |
| v2 | |
| weight |
| void Graph::edgeLabelsVisibilitySet | ( | const bool & | toggle | ) |
Toggles the visibility of edge labels.
| toggle |
| void Graph::edgeOutboundStatusSet | ( | const int & | source, |
| const int & | target, | ||
| const bool & | toggle = false |
||
| ) |
Toggles the status of outbound edge source -> target at source vertex.
| v1 | |
| v2 | |
| toggle |
| void Graph::edgeRemove | ( | const int & | v1, |
| const int & | v2, | ||
| const bool & | removeReverse = false |
||
| ) |
Removes the directed arc v1->v2 or, if the graph is undirected, the edge v1 <->v2.
Emits signal to GW to delete the graphics item.
| v1 | |
| v2 | |
| removeReverse | if true also removes the reverse edge |
| void Graph::edgeRemoveSelected | ( | SelectedEdge & | selectedEdge, |
| const bool & | removeReverse | ||
| ) |
Removes a SelectedEdge.
| selectedEdge | |
| removeReverse |
| void Graph::edgeRemoveSelectedAll | ( | ) |
Removes all selected edges.
| int Graph::edgesEnabled | ( | ) |
Returns the number |E| of graph - only the enabled edges.
| bool Graph::edgeSymmetric | ( | const int & | v1, |
| const int & | v2 | ||
| ) |
Returns TRUE if edge(v1, v2) is symmetric, i.e. (v1,v2) == (v2,v1).
| v1 | |
| v2 |
| void Graph::edgeTypeSet | ( | const int & | v1, |
| const int & | v2, | ||
| const qreal & | weight, | ||
| const int & | dirType = EdgeType::Directed |
||
| ) |
Changes the direction type of an existing edge.
| v1 | |
| v2 | |
| weight |
| qreal Graph::edgeWeight | ( | const int & | v1, |
| const int & | v2 | ||
| ) | const |
Returns the weight of the edge v1->v2.
| v1 | |
| v2 |
| void Graph::edgeWeightNumbersVisibilitySet | ( | const bool & | toggle | ) |
Changes the visibility of edge weight numbers.
| toggle |
| void Graph::edgeWeightSet | ( | const int & | v1, |
| const int & | v2, | ||
| const qreal & | weight, | ||
| const bool & | undirected = false |
||
| ) |
Changes the weight of the edge from vertex v1 to v2 (and optionally of the reverse edge)
| v1 | |
| v2 | |
| weight | |
| undirected |
| int Graph::factorial | ( | int | x | ) |
Calculates and returns x! factorial... used in (n 2)p edges calculation
| int Graph::getFileFormat | ( | ) | const |
Returns the format of the last file opened.
| QString Graph::getFileName | ( | ) | const |
Returns the file name of the current graph, if any.
| int Graph::getGeodesicsCount | ( | ) |
Returns the number of geodesics (shortest-paths) in the graph.
| QString Graph::getName | ( | ) | const |
Returns the name of the current graph.
If graph name is empty, then returns current relation name. If no relation exists, returns "noname"
| int Graph::getProminenceIndexByName | ( | const QString & | prominenceIndexName | ) |
Returns the IndexType of the given prominence index name Called from MW::slotEditNodeFind, MW::slotLayoutRadialByProminenceIndex etc.
| prominenceIndexName |
| QList< SelectedEdge > Graph::getSelectedEdges | ( | ) | const |
Returns a QList of user-selected edges in pair<int,int>
| int Graph::getSelectedEdgesCount | ( | ) | const |
Returns the count of user-selected edges.
| QList< int > Graph::getSelectedVertices | ( | ) | const |
Returns a QList of user-selected vertices.
| int Graph::getSelectedVerticesCount | ( | ) | const |
Returns count of user-selected vertices.
| int Graph::getSelectedVerticesMax | ( | ) | const |
Returns max of user-selected vertices.
| int Graph::getSelectedVerticesMin | ( | ) | const |
Returns min of user-selected vertices.
|
slot |
Resets the clicked edge and node.
Usually, called when the user clicks on an empty space.
| p |
| void Graph::graphCliqueAdd | ( | const QList< int > & | clique | ) |
Called from Graph::graphCliques to add a new clique (list of vertices) Adds clique info to each clique member and updates co-membership matrix CLQM .
| list |
| void Graph::graphCliques | ( | QSet< int > | R = QSet<int>(), |
| QSet< int > | P = QSet<int>(), |
||
| QSet< int > | X = QSet<int>() |
||
| ) |
Finds all maximal cliques in an undirected (?) graph. Implements the Bron–Kerbosch algorithm, a recursive backtracking algorithm that searches for all maximal cliques in a given graph G. Given three sets R, P, and X, the algorithm finds the maximal cliques that include all of the vertices in R, some of the vertices in P, and none of the vertices in X. In each call to the algorithm, P and X are disjoint sets whose union consists of those vertices that form cliques when added to R. In other words, P ∪ X is the set of vertices which are joined to every element of R. When P and X are both empty there are no further elements that can be added to R, so R is a maximal clique and the algorithm outputs R. The recursion is initiated by setting R and X to be the empty set and P to be the vertex set of the graph. Within each recursive call, the algorithm considers the vertices in P in turn. if there are no vertices, it either reports R as a maximal clique (if X is empty), or backtracks. For each vertex v chosen from P, it makes a recursive call in which v is added to R and in which P and X are restricted to the neighbor set NBS(v) of v, which finds and reports all clique extensions of R that contain v. Then, it moves v from P to X to exclude it from consideration in future cliques and continues with the next vertex in P.
| R | |
| P | |
| X |
| int Graph::graphCliquesContaining | ( | const int & | actor, |
| const int & | size = 0 |
||
| ) |
Returns the number of maximal cliques which include a given actor
| int Graph::graphCliquesOfSize | ( | const int & | size | ) |
Graph::graphCliquesOfSize Returns the number of maximal cliques of a given size.
| size |
| bool Graph::graphClusteringHierarchical | ( | Matrix & | STR_EQUIV, |
| const QString & | varLocation, | ||
| const int & | metric, | ||
| const int & | method, | ||
| const bool & | diagonal = false, |
||
| const bool & | diagram = false, |
||
| const bool & | considerWeights = true, |
||
| const bool & | inverseWeights = false, |
||
| const bool & | dropIsolates = false |
||
| ) |
Performs an hierarchical clustering process (Johnson, 1967) on a given NxN distance/dissimilarity matrix. The input matrix can be the the adjacency matrix, the geodesic distance matrix or a derived from them dissimilarities matrix using a user-specified metric, i.e. euclidean distance. The method parameter defines how to compute distances (similarities) between a new cluster the old clusters. Valid values can be:
| matrix | |
| metric | |
| method | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| int Graph::graphClusteringMethodStrToType | ( | const QString & | method | ) | const |
Helper method, return clustering method type from the human readable name of it.
| method |
| QString Graph::graphClusteringMethodTypeToString | ( | const int & | methodType | ) | const |
Helper method, return the human readable name of clustering method type.
| int Graph::graphConnectednessFull | ( | const bool | updateProgress = false | ) |
| qreal Graph::graphDensity | ( | ) |
Gets the graph density (if computed) or computes it again.
The graph density is the ratio of present edges to total possible edges in the current relation.
| int Graph::graphDiameter | ( | const bool | considerWeights, |
| const bool | inverseWeights | ||
| ) |
Returns the diameter of the graph, aka the largest geodesic distance between any two vertices.
| considerWeights | |
| inverseWeights |
| void Graph::graphDichotomization | ( | const qreal | threshold | ) |
Creates a new binary relation in a valued network using edge dichotomization according to the threshold parameter.
| threshold |
| qreal Graph::graphDistanceEuclidean | ( | const QPointF & | a | ) |
the euclideian distance of QPointF a (where a is difference vector)
| a |
| qreal Graph::graphDistanceEuclidean | ( | const QPointF & | a, |
| const QPointF & | b | ||
| ) |
Computes the euclideian distance between QPointF a and b.
| a | |
| b |
| int Graph::graphDistanceGeodesic | ( | const int & | v1, |
| const int & | v2, | ||
| const bool & | considerWeights = false, |
||
| const bool & | inverseWeights = true |
||
| ) |
Returns the geodesic distance (length of shortest path) from vertex v1 to vertex v2.
| v1 | |
| v2 | |
| considerWeights | |
| inverseWeights |
| qreal Graph::graphDistanceGeodesicAverage | ( | const bool | considerWeights, |
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Returns the average distance of the graph.
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::graphDistancesGeodesic | ( | const bool & | computeCentralities = false, |
| const bool & | considerWeights = false, |
||
| const bool & | inverseWeights = true, |
||
| const bool & | dropIsolates = false |
||
| ) |
Computes the geodesic distances between all vertices: In the process, it also computes many other centrality/prestige metrics:
| centralities | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
|
slot |
Stores loaded file name, graph name, sets edge direction type and signals MW to update the UI.
Called from Parser when file parsing ends.
| fileType | |
| fileName | |
| netName | |
| totalNodes | |
| totalLinks | |
| edgeDirType | |
| elapsedTime | |
| message |
|
inline |
|
slot |
Graph::graphLoadedTerminateParserThreads.
| reason |
| void Graph::graphMatrixDistanceGeodesicCreate | ( | const bool & | considerWeights = false, |
| const bool & | inverseWeights = true, |
||
| const bool & | dropIsolates = false |
||
| ) |
Creates the matrix DM of geodesic distances between vertices.
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::graphMatrixShortestPathsCreate | ( | const bool & | considerWeights = false, |
| const bool & | inverseWeights = true, |
||
| const bool & | dropIsolates = false |
||
| ) |
Creates the matrix SIGMA of shortest paths (geodesics) between vertices Each SIGMA(i,j) is the number of shortest paths (geodesics) from i and j.
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| int Graph::graphMatrixStrToType | ( | const QString & | matrix | ) | const |
Helper method, return the matrix type of human readable matrix name .
| matrix |
| QString Graph::graphMatrixTypeToString | ( | const int & | matrixType | ) | const |
Helper method, return the human readable name of matrix type.
| matrix |
| int Graph::graphMetricStrToType | ( | const QString & | metricStr | ) | const |
Helper method, return the identifier of a metric.
| metricStr |
| QString Graph::graphMetricTypeToString | ( | const int & | metricType | ) | const |
Helper method, return the human readable name of metric type.
| metric |
| bool Graph::graphReachable | ( | const int & | v1, |
| const int & | v2 | ||
| ) |
Returns true if vertices v1 and v2 are reachable.
| v1 | |
| v2 |
| qreal Graph::graphReciprocity | ( | ) |
Gets the arc reciprocity of the graph.
Also computes the dyad reciprocity and fills parameters with values.
| bool Graph::graphTriadCensus | ( | ) |
Graph::graphTriadCensus Conducts a triad census and updates QList::triadTypeFreqs, which is the list carrying all triad type frequencies Complexity:O(n!)
| void Graph::graphWalksMatrixCreate | ( | const int & | N = 0, |
| const int & | length = 0, |
||
| const bool & | updateProgress = false |
||
| ) |
Computes either the "Walks of given length" or the "Total Walks" matrix. If length>0, it computes the Walks of given length matrix, XM=AM^l where each element (i,j) denotes the number of walks of length l between vertex i and j. If length=0, it computes the Total Walks matrix, XSM=Sum{AM^n} where each (i,j) denotes the total number of walks of any length between vertices i and j. NOTE: In the latter case, this function is VERY SLOW on large networks (n>50), since it will calculate all powers of the sociomatrix up to n-1 in order to find out all possible walks.
| length | |
| updateProgress |
|
slot |
Helper method, returns a nice qstring where all html special chars are encoded.
| str |
| bool Graph::isConnected | ( | ) |
Checks if the graph is connected, in the sense of a topological space, i.e., there is a path from any vertex to any other vertex in the graph. Called from MW::slotConnectedness()
| bool Graph::isDirected | ( | ) |
Returns true if graph is directed.
| bool Graph::isEmpty | ( | ) | const |
Returns true if the current graph has no vertices at all.
| bool Graph::isFileFormatExportSupported | ( | const int & | fileFormat | ) | const |
Returns true if the fileFormat is supported for saving.
| fileFormat |
| bool Graph::isLoaded | ( | ) | const |
Returns true if a graph has been loaded from a file.
| bool Graph::isModified | ( | ) | const |
Returns true of graph is modified (edges/vertices added/removed)
| bool Graph::isSaved | ( | ) | const |
Returns true if the graph is saved.
| bool Graph::isSymmetric | ( | ) |
Returns TRUE if the adjacency matrix of the current relation is symmetric.
| bool Graph::isUndirected | ( | ) |
Returns true if graph is undirected.
| bool Graph::isWeighted | ( | ) |
Returns true if the graph is weighted (valued), i.e. if any e in |E| has value not 0 or 1 Complexity: O(n^2)
| void Graph::layoutByProminenceIndex | ( | int | prominenceIndex, |
| int | layoutType, | ||
| const bool & | considerWeights = false, |
||
| const bool & | inverseWeights = false, |
||
| const bool & | dropIsolates = false |
||
| ) |
Applies a layout according to each actor's prominence index score. The layout type can be radial (0), level (1), node sizes (2) or node colors (3), as follows: layoutType=0 - Repositions all nodes on the periphery of concentric circles with radius analogous to their prominence index layoutType=1 - Repositions all nodes on different top-down levels according to their centrality layoutType=2 - Changes node sizes to be proportional to their prominence index score layoutType=2 - Changes node colors to reflect their prominence index score (from red to green)
| prominenceIndex,0-12 | |
| layoutType | 0,1,2,3 |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::layoutCircular | ( | const double & | x0, |
| const double & | y0, | ||
| const double & | newRadius, | ||
| const bool & | guides = false |
||
| ) |
Repositions all nodes on the periphery of a circle with given radius.
| x0 | |
| y0 | |
| maxRadius |
| void Graph::layoutForceDirected_Eades_moveNodes | ( | const qreal & | c4 | ) |
| qreal Graph::layoutForceDirected_F_att | ( | const QString | model, |
| const qreal & | dist, | ||
| const qreal & | optimalDistance | ||
| ) |
| qreal Graph::layoutForceDirected_F_rep | ( | const QString | model, |
| const qreal & | dist, | ||
| const qreal & | optimalDistance | ||
| ) |
| void Graph::layoutForceDirected_FR_moveNodes | ( | const qreal & | temperature | ) |
Graph::layoutForceDirected_FR_moveNodes.
| temperature |
| qreal Graph::layoutForceDirected_FR_temperature | ( | const int | iteration | ) | const |
Reduces the temperature as the layout approaches a better configuration.
| void Graph::layoutForceDirectedFruchtermanReingold | ( | const int | maxIterations | ) |
Embeds a Force Directed Placement layout according to the Fruchterman-Reingold model. Fruchterman and Reingold (1991) refined the Spring Embedder model by replacing the forces. In this model, "the vertices behave as atomic particles or celestial bodies, exerting attractive and repulsive forces on one another." (ibid). Again, only vertices that are neighbours attract each other but, unlike Spring Embedder, all vertices repel each other. These forces induce movement. The algorithm might resemble molecular or planetary simulations, sometimes called n-body problems.
| maxIterations |
| void Graph::layoutForceDirectedKamadaKawai | ( | const int | maxIterations = 500, |
| const bool | considerWeights = false, |
||
| const bool | inverseWeights = false, |
||
| const bool | dropIsolates = false, |
||
| const QString & | initialPositions = "current" |
||
| ) |
Embeds a Force Directed Placement layout according to the Kamada-Kawai model. In this model, the network is considered to be a dynamic system where every two actors are 'particles' mutually connected by a 'spring'. Each spring has a desirable length, which corresponds to their graph theoretic distance. In this way, the optimal layout of the graph is the state with the minimum imbalance. The degree of imbalance is formulated as the total spring energy: the square summation of the differences between desirable distances and real ones for all pairs of particles Initially, the particles/actors are placed on the vertices of a regular n-polygon.
| void Graph::layoutForceDirectedSpringEmbedder | ( | const int | maxIterations | ) |
Embeds a Force Directed Placement layout according to the initial Spring Embedder model proposed by Eades.
| maxIterations | The Spring Embedder model (Eades, 1984), part of the Force Directed Placement (FDP) family, assigns forces to all vertices and edges, as if nodes were electrically charged particles (Coulomb's law) and all edges were springs (i.e. Hooke's law). |
These forces are applied to the nodes iteratively, pulling them closer together or pushing them further apart, until the system comes to an equilibrium state (node positions do not change anymore).
Note that, following Eades, we do not need to have a faithful simulation; we can -and we do- apply unrealistic forces in an unrealistic manner. For instance, instead of the forces described by Hooke's law, we will assume weaker logarithmic forces between far apart vertices...
compute max spring length as function of canvas area divided by the total vertices area
calculate electric (repulsive) forces between all vertices.
calculate spring forces between adjacent nodes that pull them together (if d > naturalLength) or push them apart (if d < naturalLength)
| void Graph::layoutRadialRandom | ( | const bool & | guides = true | ) |
Repositions all nodes on the periphery of different circles with random radius.
| x0 | |
| y0 | |
| maxRadius |
| void Graph::layoutRandom | ( | ) |
Repositions all nodes on random positions Emits setNodePos(i, x,y) to tell GW that the node item should be moved.
| maxWidth | |
| maxHeight |
| void Graph::layoutVertexSizeByIndegree | ( | ) |
Convenience method Changes the size of all nodes to be proportional to their InDegree (Degree Prestige) Calls layoutByProminenceIndex.
| void Graph::layoutVertexSizeByOutdegree | ( | ) |
Convenience method Changes the size of all nodes to be proportional to their outDegree (Degree Centrality) Calls layoutByProminenceIndex.
| void Graph::loadFile | ( | const QString | fileName, |
| const QString | codecName, | ||
| const int | fileFormat, | ||
| const QString | delimiter = QString(), |
||
| const int | sm_two_mode = 1, |
||
| const bool | sm_has_labels = false |
||
| ) |
Loads a graph from a given file.
It creates a new Parser object, moves it to a another thread, connects signals and slots and calls its run() method.
| fileName | |
| codecName | |
| m_showLabels | |
| maxWidth | |
| maxHeight | |
| fileFormat | |
| sm_two_mode |
|
private |
Computes minimum and maximum centralities during graphDistancesGeodesic()
| C | |
| v | |
| max | |
| min | |
| maxNode | |
| minNode |
| qreal Graph::numberOfTriples | ( | int | v1 | ) |
Returns the number of triples of vertex v1 A triple Υ at a vertex v is a path of length two for which v is the center vertex.
| void Graph::prestigeDegree | ( | const bool & | considerWeights, |
| const bool & | dropIsolates = false |
||
| ) |
Computes the Degree Prestige (in-degree) of each vertex - diagonal included Also the mean value and the variance of the in-degrees.
| weights | |
| dropIsolates |
| void Graph::prestigePageRank | ( | const bool & | dropIsolates = false | ) |
Calculates the PageRank Prestige of each vertex.
| dropIsolates |
| void Graph::prestigeProximity | ( | const bool | considerWeights = false, |
| const bool | inverseWeights = false, |
||
| const bool | dropIsolates = false |
||
| ) |
Computes Proximity Prestige of each vertex Also the mean value and the variance of it..
| void Graph::prominenceDistribution | ( | const int & | index, |
| const ChartType & | type, | ||
| const QString & | distImageFileName = QString() |
||
| ) |
Computes the distribution of a centrality index score. The distribution is stored as Qt Series depending on the SeriesType parameter type It is send to MW through signal/slot.
| index | |
| type |
| void Graph::prominenceDistributionArea | ( | const H_StrToInt & | discreteClasses, |
| const QString & | name, | ||
| const QString & | distImageFileName | ||
| ) |
Computes the distribution of a centrality index scores. The distribution data are returned as QAreaSeries series to MW which in turn displays them on a Area Chart.
| index | |
| series |
| void Graph::prominenceDistributionBars | ( | const H_StrToInt & | discreteClasses, |
| const QString & | name, | ||
| const QString & | distImageFileName | ||
| ) |
Computes the distribution of a centrality index scores. The distribution data are returned as QBarSeries series (with a QBarSet attached) to MW which in turn displays them on a Bar Chart.
| index | |
| series | |
| set | |
| strX |
| void Graph::prominenceDistributionSpline | ( | const H_StrToInt & | discreteClasses, |
| const QString & | seriesName, | ||
| const QString & | distImageFileName | ||
| ) |
Computes the distribution of a centrality index scores. The distribution data are returned as QSplineSeries series to MW which in turn displays them on a Spline Chart.
| index | |
| series |
| void Graph::randomizeThings | ( | ) |
Adds a little universal randomness :)
RANDOM NETWORKS
| void Graph::randomNetErdosCreate | ( | const int & | N, |
| const QString & | model, | ||
| const int & | m, | ||
| const qreal & | p, | ||
| const QString & | mode, | ||
| const bool & | diag | ||
| ) |
Creates an erdos-renyi random network according to the given model.
| vert | |
| model | |
| edges | |
| eprob | |
| mode | |
| diag |
| void Graph::randomNetLatticeCreate | ( | const int & | N, |
| const int & | length, | ||
| const int & | dimension, | ||
| const int & | neighborhoodLength, | ||
| const QString & | mode, | ||
| const bool & | circular = false |
||
| ) |
Creates a lattice network.
| N | |
| length | |
| dimension | |
| neighborhood | |
| mode | |
| diag |
| void Graph::randomNetRegularCreate | ( | const int & | N, |
| const int & | degree, | ||
| const QString & | mode, | ||
| const bool & | diag | ||
| ) |
Creates a random network where nodes have the same degree.
| vert | |
| degree |
| void Graph::randomNetRingLatticeCreate | ( | const int & | N, |
| const int & | degree, | ||
| const bool | updateProgress = false |
||
| ) |
Creates a random ring lattice network.
| vert | |
| degree | |
| x0 | |
| y0 | |
| radius | |
| updateProgress |
| void Graph::randomNetScaleFreeCreate | ( | const int & | N, |
| const int & | power, | ||
| const int & | m0, | ||
| const int & | m, | ||
| const qreal & | alpha, | ||
| const QString & | mode | ||
| ) |
Creates a scale-free random-network network.
| N | |
| power | |
| m0 | |
| m | |
| alpha | |
| mode |
| void Graph::randomNetSmallWorldCreate | ( | const int & | N, |
| const int & | degree, | ||
| const double & | beta, | ||
| const QString & | mode | ||
| ) |
Creates a small world random network.
| vert | |
| degree | |
| beta |
| void Graph::relationAdd | ( | const QString & | relName, |
| const bool & | changeRelation = false |
||
| ) |
| void Graph::relationAddCocitation | ( | ) |
Creates a new symmetric relation by connecting those actors that are cocitated by others. In the new relation, an edge will exist between actor i and actor j only if C(i,j) > 0, where C the Cocitation Matrix. Thus the actor pairs cited by more common neighbors will appear with a stronger tie between them than pairs those cited by fewer common neighbors. The resulting relation is symmetric.
|
slot |
Gets the current relation number.
|
slot |
Gets the current relation name.
|
slot |
Overload. Renames current relation to newName, without emitting any signal to MW.
| newName |
|
slot |
Renames current relation to newName, optionally emitting a signal to MW.
| newName |
|
slot |
Changes graph to next relation.
|
slot |
Changes graph to previous relation.
| int Graph::relations | ( | ) |
Returns the count of relationships in this Graph.
| void Graph::relationsClear | ( | ) |
Clears relationships in this Graph.
|
slot |
Changes the current relation, and optionally emits signals to MW/GW (default: true)
Slots to signals from MainWindow
Forces all enabled vertices to disable edges in the old relation and enable edges of the new relation
Then, if updateUI==true (default), it emits signals to MW and GW to update the MW UI and toggle the edges on the GW, respectivelly.
Called from Parser, Graph methods and when the user selects a relation in the MW combo box.
| relNum | int |
| updateUI | bool |
|
private |
Checks if score C is a new prominence class If yes, it stores that number in a QHash<QString,int> type where the score is the key. If no, increases the frequency of this prominence score by 1 Called from graphDistancesGeodesic()
| C | |
| discreteClasses | |
| classes |
|
private |
Overloaded method. It only adds displaying current vertex for debugging purposes.
| C | |
| discreteClasses | |
| classes | |
| vertex |
| bool Graph::saveToAdjacencyFormat | ( | const QString & | fileName, |
| const bool & | saveEdgeWeights = true |
||
| ) |
Saves the active graph to an adjacency-formatted file.
| fileName |
| bool Graph::saveToDotFormat | ( | QString | fileName | ) |
TODO Saves the active graph to a UCINET-formatted file.
| fileName |
| void Graph::saveToFile | ( | const QString & | fileName, |
| const int & | fileType, | ||
| const bool & | saveEdgeWeights = true, |
||
| const bool & | saveZeroWeightEdges = false |
||
| ) |
Saves the current graph to a file.
Checks the requested file type and calls the corresponding saveGraphTo...() method
| fileName | |
| fileType |
| bool Graph::saveToGraphMLFormat | ( | const QString & | fileName, |
| const bool & | saveZeroWeightEdges = false, |
||
| QString | networkName = "", |
||
| int | maxWidth = 0, |
||
| int | maxHeight = 0 |
||
| ) |
Saves the current graph to a GraphML-formatted file.
| fileName | |
| networkName | |
| maxWidth | |
| maxHeight |
| bool Graph::saveToPajekFormat | ( | const QString & | fileName, |
| QString | networkName = "", |
||
| int | maxWidth = 0, |
||
| int | maxHeight = 0 |
||
| ) |
Saves the active graph to a Pajek-formatted file.
Preserves node properties (positions, colours, etc)
| fileName | |
| networkName | |
| maxWidth | |
| maxHeight |
| void Graph::setDirected | ( | const bool & | toggle = true, |
| const bool & | signalMW = true |
||
| ) |
Toggles the graph directed or undirected.
| toggle | |
| signalMW |
|
signal |
|
signal |
|
signal |
| void Graph::setFileFormat | ( | const int & | fileFormat | ) |
| void Graph::setFileName | ( | const QString & | fileName | ) |
Sets the file name of the current graph.
| fileName |
| void Graph::setModStatus | ( | const int & | graphNewStatus, |
| const bool & | signalMW = true |
||
| ) |
Sets the graph modification status.
If there are major changes or new network, it signals to MW to update the UI.
| int | graphNewStatus |
| bool | signalMW |
| void Graph::setName | ( | const QString & | graphName | ) |
Sets the name of the current graph.
| graphName |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
| void Graph::setReportsChartType | ( | const int & | type | ) |
Sets the chart type in reports.
| type |
| void Graph::setReportsDataDir | ( | const QString & | dir | ) |
Sets the directory where reports are saved This is used when exporting prominence distribution images to be used in HTML reports.
| dir |
| void Graph::setReportsLabelLength | ( | const int & | length | ) |
Sets the length of labels in reports.
| length |
| void Graph::setReportsRealNumberPrecision | ( | const int & | precision | ) |
Sets the precision (number of fraction digits) the app will use when writing real numbers in reports.
| precision |
|
slot |
Sets the user-selected vertices and edges.
Usually called from GW, it emits selection counts to MW
| selectedVertices | |
| selectedEdges |
| void Graph::setSymmetric | ( | ) |
Transforms the graph to symmetric (all edges reciprocal)
| void Graph::setUndirected | ( | const bool & | toggle = true, |
| const bool & | signalMW = true |
||
| ) |
Makes the graph undirected or directed.
| toggle | |
| signalMW |
|
signal |
| void Graph::setWeighted | ( | const bool & | toggle = true | ) |
Sets the graph to be weighted ( valued edges ).
| toggle |
| int Graph::sign | ( | const qreal & | D | ) |
|
signal |
|
signal |
|
signal |
Signals to GraphicsWidget
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
Signals to MainWindow
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
signal |
|
slot |
Gets the reply of a MW network request made by Web Crawler, and emits that reply as is to the Web Crawler.
|
slot |
Creates a new WebCrawler, that will parse the downloaded HTML code of each webpage we download. Moves the WebCrawler to a new thread and starts the thread. Then creates the fist node (initial url), and starts the web spider to download the first HTML page. Called by MW with user options.
| startUrl | |
| urlPatternsIncluded | |
| urlPatternsExcluded | |
| linkClasses | |
| maxNodes | |
| maxLinksPerPage | |
| intLinks | |
| childLinks | |
| parentLinks | |
| selfLinks | |
| extLinksIncluded | |
| extLinksCrawl | |
| socialLinks | |
| delayedRequests |
|
signal |
| void Graph::triadType_examine_MAN_label | ( | int | mut, |
| int | asy, | ||
| int | nul, | ||
| GraphVertex * | vert1, | ||
| GraphVertex * | vert2, | ||
| GraphVertex * | vert3 | ||
| ) |
Examines the triad type (in Mutual-Asymmetric-Null label format) and increases by one the proper frequency element inside QList::triadTypeFreqs
| int Graph::vertexClicked | ( | ) | const |
Returns the number of the clicked vertex.
|
slot |
Sets the clicked vertex.
Signals to MW to show node info on the status bar.
| v1 | |
| p |
| QColor Graph::vertexColor | ( | const int & | v1 | ) | const |
| void Graph::vertexColorInit | ( | const QString & | color | ) |
Graph::vertexColorInit default vertex color initialization.
| color |
| void Graph::vertexColorSet | ( | const int & | v1, |
| const QString & | color | ||
| ) |
Changes the color of vertex v1.
| v1 | |
| color |
|
slot |
Creates a new vertex.
Slots to signals from Parser
Main vertex creation slot, associated with homonymous signal from Parser. Adds a vertex to the Graph and signals drawNode to GW The new vertex has number num and specific color, label, label color, shape and position p.
| num | |
| size | |
| nodeColor | |
| numColor | |
| numSize | |
| label | |
| lColor | |
| lSize | |
| p | |
| nodeShape | |
| signalMW |
|
slot |
Creates a new vertex in the given position.
Called from GW, with i and p as parameters. Calls the main creation slot with init node values.
| QPointF | The clicked pos of the new node. |
|
slot |
Creates a new randomly positioned vertex with default values.
Computes a random position p inside the useable canvas area Then calls the main creation slot with init node values.
| bool |
|
slot |
Creates a new randomly positioned vertex with specific number and label. All other values are from the defaults.
Called from WebCrawler and Parser with parameters label and i. Computes a random position p the useable canvas area Then calls the main creation slot with init node values.
| i | |
| label | |
| signalMW |
| int Graph::vertexDegreeIn | ( | int | v1 | ) |
Returns the inDegree (sum of inbound edge weights) of vertex v1.
| v1 |
| int Graph::vertexDegreeOut | ( | int | v1 | ) |
Returns the outDegree (sum of outbound edge weights) of vertex v1.
| v1 |
| int Graph::vertexEdgesInbound | ( | int | v1 | ) |
Returns the number of inbound edges (arcs) to vertex v1.
| v1 |
| int Graph::vertexEdgesOutbound | ( | int | v1 | ) |
Returns the number of outbound edges (arcs) from vertex v1.
| v1 |
| int Graph::vertexExists | ( | const int & | v1 | ) |
Checks if the given vertex exists in the graph.
Returns the vpos or -1
Complexity: O(logN) for vpos retrieval
| vertex | number |
| int Graph::vertexExists | ( | const QString & | label | ) |
Checks if there is a vertex with a specific label exists in the graph.
Returns the vpos or -1
Complexity: O(N)
| label |
| bool Graph::vertexFindByIndexScore | ( | const int & | index, |
| const QStringList & | thresholds, | ||
| const bool & | considerWeights, | ||
| const bool & | inverseWeights = false, |
||
| const bool & | dropIsolates = false |
||
| ) |
Finds vertices by their index score.
| QStringList |
| bool Graph::vertexFindByLabel | ( | const QStringList & | labelList | ) |
Finds vertices by their label.
| QStringList |
| bool Graph::vertexFindByNumber | ( | const QStringList & | numList | ) |
Finds vertices in strList by their number.
| QStringList |
| QList< int > Graph::vertexinfluenceDomain | ( | int | v1 | ) |
Returns the influence domain of vertex v1, namely the set of nodes who can reach v1 The Influence Domain Ii of vertex v can also be defined as: Ii = Sum [ D(i,v), iff D(i,v) != inf ] for every in V, where i!=v and D the distance matrix This function applies to digraphs only.
| v1 |
| QList< int > Graph::vertexinfluenceRange | ( | int | v1 | ) |
Returns the influence range of vertex v1, namely the set of nodes who are reachable from v1 (See Wasserman and Faust, pp.200-201, based on Lin, 1976). The Influence Range of vertex v can also be defined as: Ji = Sum [ D(v,j), iff D(v,j) != inf ] for every j in V, where j!=v and D the distance matrix This function is for digraphs only.
| v1 |
| bool Graph::vertexIsolated | ( | const int & | v1 | ) | const |
Checks if vertex is isolated.
| v1 |
|
slot |
Toggles the status of all isolated vertices (thos without links)
For each isolate vertex in the Graph, emits the setVertexVisibility signal
| toggle |
| QString Graph::vertexLabel | ( | const int & | v | ) | const |
Returns the label of a vertex v1.
| v1 |
| void Graph::vertexLabelColorInit | ( | QString | color | ) |
Graph::vertexLabelColorInit Changes the default vertex label color.
| color |
| void Graph::vertexLabelColorSet | ( | const int & | v1, |
| const QString & | color | ||
| ) |
Changes the label color of vertex v1 or all vertices if v1 = 0.
| v1 | |
| color |
| void Graph::vertexLabelDistanceAllSet | ( | const int & | newDistance | ) |
Changes the distance of all vertex labels from their vertices.
| size |
| void Graph::vertexLabelDistanceInit | ( | const int & | distance | ) |
Changes the default distance of vertex labels.
| distance |
| void Graph::vertexLabelDistanceSet | ( | const int & | v, |
| const int & | newDistance | ||
| ) |
Changes the distance.of vertex v label from the vertex.
| v | |
| size |
| void Graph::vertexLabelSet | ( | const int & | v1, |
| const QString & | label | ||
| ) |
Changes the label of a vertex v1.
| v1 | |
| label |
| void Graph::vertexLabelSizeInit | ( | int | newSize | ) |
Graph::vertexLabelSizeInit Changes the default size of vertex labels.
| newSize |
| void Graph::vertexLabelSizeSet | ( | const int & | v1, |
| const int & | labelSize | ||
| ) |
Changes the label size of vertex v1 or all vertices if v1=0.
| v1 | |
| size |
| void Graph::vertexLabelsVisibilitySet | ( | bool | toggle | ) |
| QList< int > Graph::vertexNeighborhoodList | ( | const int & | v1 | ) |
Returns a list of all vertices mutually connected to vertex v1 in the current relation.
| v1 |
| QSet< int > Graph::vertexNeighborhoodSet | ( | const int & | v1 | ) |
Returns the set of all vertices mutually connected to vertex v1 in the current relation.
| v1 |
| void Graph::vertexNumberColorInit | ( | const QString & | color | ) |
Changes the initial color of the vertex numbers.
| color |
| void Graph::vertexNumberColorSet | ( | const int & | v1 = 0, |
| const QString & | color = "#000000" |
||
| ) |
Graph::vertexColorSet Changes the color of vertex v1.
| v1 | |
| color |
| void Graph::vertexNumberDistanceInit | ( | const int & | distance | ) |
Changes the initial distance of vertex numbers.
| distance |
| void Graph::vertexNumberDistanceSet | ( | const int & | v, |
| const int & | newDistance | ||
| ) |
Changes the distance.of vertex v number from the vertex.
| v | |
| size |
| int Graph::vertexNumberMax | ( | ) |
Returns the number of the last vertex in the graph.
| int Graph::vertexNumberMin | ( | ) |
Returns the number of the first vertex in the graph.
| void Graph::vertexNumberSizeInit | ( | const int & | size | ) |
Changes the initial size of vertex numbers.
| size |
| void Graph::vertexNumberSizeSet | ( | const int & | v, |
| const int & | size | ||
| ) |
Changes the size of vertex v number.
| v | |
| size |
| QPointF Graph::vertexPos | ( | const int & | v1 | ) | const |
Returns the position of the given vertex.
| v1 |
| void Graph::vertexPosSet | ( | const int & | v1, |
| const int & | x, | ||
| const int & | y | ||
| ) |
Changes the position of the given vertex.
Called from MW/GW when node moves to update its position
| v1 | |
| x | |
| y |
| void Graph::vertexRemove | ( | const int & | v1 | ) |
Removes the vertex v1 from the graph First, it removes all edges to doomed from other vertices Then it changes the vpos of all subsequent vertices inside m_graph Finally, it removes the vertex.
| int | v1 |
|
slot |
Deletes a dummy node.
This is called from Parser (as pajek) to delete any redundant (dummy) nodes.
| int | i number of node |
| QString Graph::vertexShape | ( | const int & | v1 | ) |
Returns the shape of this vertex.
| v1 |
| QString Graph::vertexShapeIconPath | ( | const int & | v1 | ) |
Returns the IconPath of vertex v1.
| v1 |
| void Graph::vertexShapeSet | ( | const int & | v1, |
| const QString & | shape, | ||
| const QString & | iconPath = QString() |
||
| ) |
Changes the shape and iconPath of vertex v1, or all vertices if v1=-1.
| v1 | |
| shape | |
| iconPath |
| void Graph::vertexShapeSetDefault | ( | const QString | shape, |
| const QString & | iconPath = QString() |
||
| ) |
Sets the default vertex shape and iconPath.
| shape | |
| iconPath |
| int Graph::vertexSize | ( | const int & | v | ) | const |
Returns the size of vertex v.
| v |
| void Graph::vertexSizeInit | ( | const int | size | ) |
Sets the initial vertex size.
| size |
| void Graph::vertexSizeSet | ( | const int & | v, |
| const int & | size | ||
| ) |
Changes the size of a vertex v or all vertices if v=0.
Called from MW (i.e. user changing node properties)
| v | |
| size |
| int Graph::vertices | ( | const bool & | dropIsolates = false, |
| const bool & | countAll = false, |
||
| const bool & | recount = false |
||
| ) |
Gets the number of vertices in the graph.
If countAll = true, returns |V| where V the set of all (enabled or not) vertices If countAll = false, it skips disabled vertices If countAll = false and dropIsolates = true, it skips both disabled and isolated vertices
| dropIsolates | |
| countAll |
| void Graph::verticesCreateSubgraph | ( | QList< int > | vList, |
| const int & | type = SUBGRAPH_CLIQUE, |
||
| const int & | center = 0 |
||
| ) |
Creates a subgraph (clique, star, cycle, line) with vertices in vList Iff vList is empty, then fallbacks to the m_verticesSelected.
| vList |
| QList< int > Graph::verticesList | ( | ) |
Returns a list of all vertices numbers inside the graph.
| QList< int > Graph::verticesListIsolated | ( | ) |
Returns a list of all isolated vertices inside the graph.
| QSet< int > Graph::verticesSet | ( | ) |
Returns a QSet of all vertices numbers inside the graph.
| int Graph::verticesWithInboundEdges | ( | ) |
Returns the sum of vertices having edgesInbound.
| int Graph::verticesWithOutboundEdges | ( | ) |
Returns the sum of vertices having edgesOutbound.
| int Graph::verticesWithReciprocalEdges | ( | ) |
Returns the sum of vertices having reciprocal edges.
| int Graph::walksBetween | ( | int | v1, |
| int | v2, | ||
| int | length | ||
| ) |
Calculates and returns the number of walks of a given length between v1 and v2.
| v1 | |
| v2 | |
| length |
| void Graph::webCrawlTerminateThreads | ( | QString | reason | ) |
called from Graph, when closing network, to terminate all crawler processes Also called indirectly when wc_spider finishes
| reason |
|
slot |
A loop, that takes urls awaiting in front of the urlQueue, and signals to the MW to make the network request.
| void Graph::writeCentralityBetweenness | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes Betweenness centralities to file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeCentralityCloseness | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes the closeness centralities to a file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeCentralityClosenessInfluenceRange | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes the "improved" closeness centrality indices to a file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeCentralityDegree | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes the Degree Centrality to a file.
| fileName | |
| considerWeights | |
| dropIsolates |
| void Graph::writeCentralityEccentricity | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes the Eccentricity centralities (aka Harary Graph Centrality) to a file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeCentralityEigenvector | ( | const QString | fileName, |
| const bool & | considerWeights = true, |
||
| const bool & | inverseWeights = false, |
||
| const bool & | dropIsolates = false |
||
| ) |
Writes the eigenvector centralities to a file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeCentralityInformation | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights | ||
| ) |
Writes the information centralities to file.
| fileName | |
| considerWeights | |
| inverseWeights |
| void Graph::writeCentralityPower | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes Power Centralities to a file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeCentralityStress | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes the Stress centralities to a file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| bool Graph::writeCliqueCensus | ( | const QString & | fileName, |
| const bool | considerWeights | ||
| ) |
Graph::writeCliqueCensus Calls graphCliques() to compute all cliques (maximal connected subgraphs) of the network. Then writes the results into a file, along with the Actor by clique analysis, the Co-membership matrix and the Hierarchical clustering of overlap matrix.
| fileName | |
| considerWeights |
Write the actor by clique analysis matrix. For each actor-clique pair, we compute the proportion of clique members adjacent
Write the actor by actor analysis matrix. For each pair, we compute their clique co-membership
Write the Hierarchical clustering of overlap matrix
| void Graph::writeClusteringCoefficient | ( | const QString | fileName, |
| const bool | considerWeights | ||
| ) |
Graph::writeClusteringCoefficient Writes the clustering coefficients to a file.
| fileName | |
| considerWeights |
| bool Graph::writeClusteringHierarchical | ( | const QString & | fileName, |
| const QString & | varLocation, | ||
| const QString & | matrix = "Adjacency", |
||
| const QString & | metric = "Manhattan", |
||
| const QString & | method = "Complete", |
||
| const bool & | diagonal = false, |
||
| const bool & | dendrogram = false, |
||
| const bool & | considerWeights = true, |
||
| const bool & | inverseWeights = false, |
||
| const bool & | dropIsolates = false |
||
| ) |
Writes Hierarchical Clustering Analysis to a given file.
| fileName | |
| matrix | |
| similarityMeasure | |
| method | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeClusteringHierarchicalResultsToStream | ( | QTextStream & | outText, |
| const int | N, | ||
| const bool & | dendrogram = false |
||
| ) |
Writes Hierarchical Clustering results to given output stream Before running this methos, the method Graph::graphClusteringHierarchical() must execute and return true. Otherwise, the result is unpredictable...
| outText | |
| N | |
| dendrogram |
| void Graph::writeDataSetToFile | ( | const QString | dir, |
| const QString | fileName | ||
| ) |
Writes a "famous" dataset to the given file Datasets are hardcoded! They are exported in the given fileName...
TODO: Move all these datasets to a separate class
| fileName |
| void Graph::writeEccentricity | ( | const QString | fileName, |
| const bool | considerWeights = false, |
||
| const bool | inverseWeights = false, |
||
| const bool | dropIsolates = false |
||
| ) |
Writes the Eccentricity report to file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeMatrix | ( | const QString & | fn, |
| const int & | matrix = MATRIX_ADJACENCY, |
||
| const bool & | considerWeights = true, |
||
| const bool & | inverseWeights = false, |
||
| const bool & | dropIsolates = false, |
||
| const QString & | varLocation = "Rows", |
||
| const bool & | simpler = false |
||
| ) |
Writes the specified matrix of social network to file fn
| void Graph::writeMatrixAdjacency | ( | const QString | fn, |
| const bool & | markDiag = true |
||
| ) |
Writes the adjacency matrix of G to a specified file fn
| void Graph::writeMatrixAdjacencyInvert | ( | const QString & | fn, |
| const QString & | method | ||
| ) |
Calls Graph::createMatrixAdjacencyInverse(method) to compute the inverse of the adjacency matrix and writes it to file fn in HTML notation.
| fn | |
| method |
| void Graph::writeMatrixAdjacencyPlot | ( | const QString | fn, |
| const bool & | simpler = false |
||
| ) |
Writes a visual representation of the adjacency matrix of the graph to the specified file.
The resulting matrix HAS NO spaces between elements.
| fn | |
| simpler |
| void Graph::writeMatrixAdjacencyTo | ( | QTextStream & | os, |
| const bool & | saveEdgeWeights = true |
||
| ) |
Exports the adjacency matrix to a given textstream
| void Graph::writeMatrixDegreeText | ( | const QString & | fn | ) |
Computes the Degree matrix of the graph and writes it to given file.
| fn |
| void Graph::writeMatrixDissimilarities | ( | const QString | fileName, |
| const QString & | metricStr, | ||
| const QString & | varLocation, | ||
| const bool & | diagonal, | ||
| const bool & | considerWeights | ||
| ) |
Writes dissimilarity matrix based on a metric/measure to given html file.
| fileName | |
| measure | |
| varLocation | |
| diagonal | |
| considerWeights |
| void Graph::writeMatrixDistancesPlainText | ( | const QString & | fn, |
| const bool & | considerWeights, | ||
| const bool & | inverseWeights, | ||
| const bool & | dropIsolates | ||
| ) |
Writes the matrix of distances to a file.
| fn | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeMatrixHTMLTable | ( | QTextStream & | outText, |
| Matrix & | M, | ||
| const bool & | markDiag = true, |
||
| const bool & | plain = false, |
||
| const bool & | printInfinity = true, |
||
| const bool & | dropIsolates = false |
||
| ) |
Writes the matrix M as HTML.
specified text stream outText It is the same as Matrix::printHTMLTable except that this method omits disabled vertices, thus the table header is correct
| outText | |
| M | |
| markDiag | |
| plain | |
| printInfinity |
| void Graph::writeMatrixLaplacianPlainText | ( | const QString & | fn | ) |
Computes the Laplacian matrix of the graph and writes it to given file.
| fn |
| void Graph::writeMatrixShortestPathsPlainText | ( | const QString & | fn, |
| const bool & | considerWeights, | ||
| const bool & | inverseWeights | ||
| ) |
Writes the shortest paths matrix to a file Each SIGMA(i,j) element is the number of shortest paths (geodesics) from i and j.
| fn | |
| considerWeights | |
| inverseWeights |
| void Graph::writeMatrixSimilarityMatching | ( | const QString | fileName, |
| const QString & | measure = "Simple", |
||
| const QString & | matrix = "adjacency", |
||
| const QString & | varLocation = "rows", |
||
| const bool & | diagonal = false, |
||
| const bool & | considerWeights = true |
||
| ) |
Writes similarity matrix based on a matching measure to given html file.
| fileName | |
| measure | |
| matrix | |
| varLocation | |
| diagonal | |
| considerWeights |
| void Graph::writeMatrixSimilarityMatchingPlain | ( | const QString | fileName, |
| const int & | measure = METRIC_SIMPLE_MATCHING, |
||
| const QString & | matrix = "adjacency", |
||
| const QString & | varLocation = "rows", |
||
| const bool & | diagonal = false, |
||
| const bool & | considerWeights = true |
||
| ) |
Writes similarity matrix based on a matching measure to given file.
| fileName | |
| measure | |
| matrix | |
| varLocation | |
| diagonal | |
| considerWeights |
| void Graph::writeMatrixSimilarityPearson | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const QString & | matrix = "adjacency", |
||
| const QString & | varLocation = "rows", |
||
| const bool & | diagonal = false |
||
| ) |
Calls Graph::createMatrixSimilarityPearson() and writes Pearson Correlation Coefficients to given file.
| fileName | |
| considerWeights |
| void Graph::writeMatrixSimilarityPearsonPlainText | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const QString & | matrix = "adjacency", |
||
| const QString & | varLocation = "rows", |
||
| const bool & | diagonal = false |
||
| ) |
Calls Graph::graphSimilariyPearsonCorrelationCoefficients() and writes Pearson Correlation Coefficients to given file.
| fileName | |
| considerWeights |
| void Graph::writeMatrixWalks | ( | const QString & | fn, |
| const int & | length = 0, |
||
| const bool & | simpler = false |
||
| ) |
Calls graphWalksMatrixCreate() to compute walks and writes the Walks of given length matrix to a file in HTML. If length = 0, it writes the Total Walks matrix.
| fn | |
| length | |
| simpler |
| void Graph::writePrestigeDegree | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes the Degree Prestige of each node to a file.
| fileName | |
| considerWeights | |
| dropIsolates |
| void Graph::writePrestigePageRank | ( | const QString | fileName, |
| const bool | dropIsolates = false |
||
| ) |
Writes the PageRank scores of vertices to a file.
| fileName | |
| dropIsolates |
| void Graph::writePrestigeProximity | ( | const QString | fileName, |
| const bool | considerWeights, | ||
| const bool | inverseWeights, | ||
| const bool | dropIsolates | ||
| ) |
Writes the proximity prestige indices to a file.
| fileName | |
| considerWeights | |
| inverseWeights | |
| dropIsolates |
| void Graph::writeReachabilityMatrixPlainText | ( | const QString & | fn, |
| const bool & | dropIsolates = false |
||
| ) |
Writes the reachability matrix X^R of the graph to a file
| void Graph::writeReciprocity | ( | const QString | fileName, |
| const bool | considerWeights = false |
||
| ) |
Writes reciprocity report to filename.
| fileName | |
| considerWeights |
| void Graph::writeTriadCensus | ( | const QString | fileName, |
| const bool | considerWeights | ||
| ) |
| void Graph::writeWalksOfLengthMatrixPlainText | ( | const QString & | fn, |
| const int & | length | ||
| ) |
Graph::writeWalksOfLengthMatrixPlainText.
| fn | |
| length |
| void Graph::writeWalksTotalMatrixPlainText | ( | const QString & | fn | ) |
Graph::writeWalksTotalMatrixPlainText Writes the total number of walks matrix.
| fn | |
| netName | |
| length |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
used in resolveClasses and graphDistancesGeodesic()
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
General & initialisation variables
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
| H_f_i Graph::sizeOfNthOrderNeighborhood |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |
| H_Int Graph::vpos |
vpos stores the real position of each vertex inside m_graph. It starts at zero (0). We need to know the place of a vertex inside m_graph after adding or removing many vertices
|
private |
|
private |
|
private |
|
private |
|
private |
|
private |